Biomolecular Assemblies: Moving from Observation to Predictive Design
- PMID: 30281290
- PMCID: PMC6650774
- DOI: 10.1021/acs.chemrev.8b00038
Biomolecular Assemblies: Moving from Observation to Predictive Design
Abstract
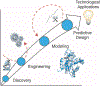
Biomolecular assembly is a key driving force in nearly all life processes, providing structure, information storage, and communication within cells and at the whole organism level. These assembly processes rely on precise interactions between functional groups on nucleic acids, proteins, carbohydrates, and small molecules, and can be fine-tuned to span a range of time, length, and complexity scales. Recognizing the power of these motifs, researchers have sought to emulate and engineer biomolecular assemblies in the laboratory, with goals ranging from modulating cellular function to the creation of new polymeric materials. In most cases, engineering efforts are inspired or informed by understanding the structure and properties of naturally occurring assemblies, which has in turn fueled the development of predictive models that enable computational design of novel assemblies. This Review will focus on selected examples of protein assemblies, highlighting the story arc from initial discovery of an assembly, through initial engineering attempts, toward the ultimate goal of predictive design. The aim of this Review is to highlight areas where significant progress has been made, as well as to outline remaining challenges, as solving these challenges will be the key that unlocks the full power of biomolecules for advances in technology and medicine.
Figures















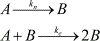

References
-
- Barabasi AL; Oltvai ZN Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet 2004, 5, 101–113. - PubMed
-
- Ross PD; Subramanian S Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 2002, 20, 3096–3102. - PubMed
-
- Luo Q; Hou C; Bai Y; Wang R; Liu J Protein Assembly: Versatile Approaches to Construct Highly Ordered Nanostructures. Chem. Rev 2016, 116, 13571–13632. - PubMed
-
- Yeates TO; Padilla JE Designing Supramolecular Protein Assemblies. Curr. Opin. Struct. Biol 2002, 12, 464–470. - PubMed
-
- Jackel C; Kast P; Hilvert D Protein Design by Directed Evolution. Annu. Rev. Biophys 2008, 37, 153–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

