Transfection of choanoflagellates illuminates their cell biology and the ancestry of animal septins
- PMID: 30281390
- PMCID: PMC6333174
- DOI: 10.1091/mbc.E18-08-0514
Transfection of choanoflagellates illuminates their cell biology and the ancestry of animal septins
Abstract
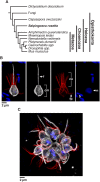
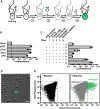
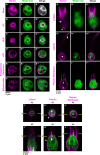
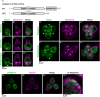
As the closest living relatives of animals, choanoflagellates offer unique insights into animal origins and core mechanisms underlying animal cell biology. However, unlike traditional model organisms, such as yeast, flies, and worms, choanoflagellates have been refractory to DNA delivery methods for expressing foreign genes. Here we report a robust method for expressing transgenes in the choanoflagellate Salpingoeca rosetta, overcoming barriers that have previously hampered DNA delivery and expression. To demonstrate how this method accelerates the study of S. rosetta cell biology, we engineered a panel of fluorescent protein markers that illuminate key features of choanoflagellate cells. We then investigated the localization of choanoflagellate septins, a family of GTP-binding cytoskeletal proteins that are hypothesized to regulate multicellular rosette development in S. rosetta. Fluorescently tagged septins localized to the basal poles of S. rosetta single cells and rosettes in a pattern resembling septin localization in animal epithelia. The establishment of transfection in S. rosetta and its application to the study of septins represent critical advances in the use of S. rosetta as an experimental model for investigating choanoflagellate cell biology, core mechanisms underlying animal cell biology, and the origin of animals.
Figures
Comment in
-
Developing Evolutionary Cell Biology.Dev Cell. 2018 Nov 19;47(4):395-396. doi: 10.1016/j.devcel.2018.11.006. Dev Cell. 2018. PMID: 30458131
References
-
- Abedin M, King N. (2008). The premetazoan ancestry of cadherins. Science , 946–948. - PubMed

