A mitochondrial ROS pathway controls matrix metalloproteinase 9 levels and invasive properties in RAS-activated cancer cells
- PMID: 30281903
- PMCID: PMC7379617
- DOI: 10.1111/febs.14671
A mitochondrial ROS pathway controls matrix metalloproteinase 9 levels and invasive properties in RAS-activated cancer cells
Abstract
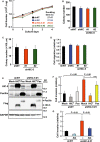
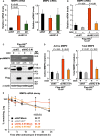
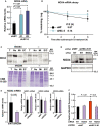
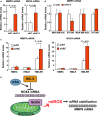
Matrix metalloproteinases (MMPs) are tissue-remodeling enzymes involved in the processing of various biological molecules. MMPs also play important roles in cancer metastasis, contributing to angiogenesis, intravasation of tumor cells, and cell migration and invasion. Accordingly, unraveling the signaling pathways controlling MMP activities could shed additional light on cancer biology. Here, we report a molecular axis, comprising the molecular adaptor hydrogen peroxide-inducible clone-5 (HIC-5), NADPH oxidase 4 (NOX4), and mitochondria-associated reactive oxygen species (mtROS), that regulates MMP9 expression and may be a target to suppress cancer metastasis. We found that this axis primarily downregulates mtROS levels which stabilize MMP9 mRNA. Specifically, HIC-5 suppressed the expression of NOX4, the source of the mtROS, thereby decreasing mtROS levels and, consequently, destabilizing MMP9 mRNA. Interestingly, among six cancer cell lines, only EJ-1 and MDA-MB-231 cells exhibited upregulation of NOX4 and MMP9 expression after shRNA-mediated HIC-5 knockdown. In these two cell lines, activating RAS mutations commonly occur, suggesting that the HIC-5-mediated suppression of NOX4 depends on RAS signaling, a hypothesis that was supported experimentally by the introduction of activated RAS into mammary epithelial cells. Notably, HIC-5 knockdown promoted lung metastasis of MDA-MB-231 cancer cells in mice. The tumor growth of HIC-5-silenced MDA-MB-231 cells at the primary sites was comparable to that of control cells. Consistently, the invasive properties of the cells, but not their proliferation, were enhanced by the HIC-5 knockdown in vitro. We conclude that NOX4-mediated mtROS signaling increases MMP9 mRNA stability and affects cancer invasiveness but not tumor growth.
Keywords: ROS; HIC-5; MMP9; NOX4; metastasis.
© 2018 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors have no conflicts of interest with the contents of this article.
Figures



References
-
- Rodriguez D, Morrison CJ & Overall CM (2010) Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics Biochem Biophys Acta 1803, 39–54. - PubMed
-
- Deryugina EI & Quigley JP (2006) Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev 25, 9–34. - PubMed
-
- Egeblad M & Werb Z (2002) New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2, 161–174. - PubMed
-
- Bjorklund M & Koivunen E (2005) Gelatinase‐mediated migration and invasion of cancer cells. Biochem Biophys Acta 1755, 37–69. - PubMed
-
- Daniele A, Abbate I, Oakley C, Casamassima P, Savino E, Casamassima A, Sciortino G, Fazio V, Gadaleta‐Caldarola G, Catino A et al (2016) Clinical and prognostic role of matrix metalloproteinase‐2, ‐9 and their inhibitors in breast cancer and liver diseases: a review. Int J Biochem Cell Biol 77, 91–101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

