Cell-lineage level-targeted sequencing to identify acute myeloid leukemia with myelodysplasia-related changes
- PMID: 30282643
- PMCID: PMC6177645
- DOI: 10.1182/bloodadvances.2017010744
Cell-lineage level-targeted sequencing to identify acute myeloid leukemia with myelodysplasia-related changes
Abstract
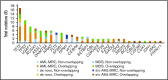
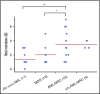
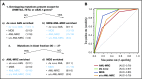
Acute myeloid leukemia (AML) is a clonal myeloid neoplasm that typically arises de novo; however, some cases evolve from a preleukemic state, such as myelodysplastic syndrome (MDS). Such secondary AMLs and those with typical MDS-related clinical features are known as AMLs with myelodysplasia-related changes (AML-MRC). Because patients with AML-MRC have poor prognosis, more accurate diagnostic approaches are required. In this study, we performed targeted sequencing of 54 genes in 3 cell populations (granulocyte, blast, and T-cell fractions) using samples from 13 patients with MDS, 16 patients with clinically diagnosed AML-MRC, 4 patients with suspected AML-MRC but clinically diagnosed as AML not otherwise specified (AML-NOS), and 11 patients with de novo AML. We found that overlapping mutations, defined as those shared at least by the blast and granulocyte fractions, were significantly enriched in patients with MDS and AML-MRC, including those with suspected AML-MRC, indicating a substantial history of clonal hematopoiesis. In contrast, blast-specific nonoverlapping mutations were significantly enriched in patients with de novo AML. Furthermore, the presence of overlapping mutations, excluding DNMT3A, TET2, and ASXL1, effectively segregated patients with MDS and AML-MRC or suspected AML-MRC from patients with de novo AML. Additionally, the presence of ≥3 mutations in the blast fraction was useful for distinguishing patients with AML-MRC from those with MDS. In conclusion, our approach is useful for classifying clinically diagnosable AML-MRC and identifying clinically diagnosed AML-NOS as latent AML-MRC. Additional prospective studies are needed to confirm the utility of this approach.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of interest disclosure: The authors declare no competing financial interests.
Figures





Similar articles
-
ASXL1 mutation as a surrogate marker in acute myeloid leukemia with myelodysplasia-related changes and normal karyotype.Cancer Med. 2020 Jun;9(11):3637-3646. doi: 10.1002/cam4.2947. Epub 2020 Mar 26. Cancer Med. 2020. PMID: 32216059 Free PMC article.
-
Acute myeloid leukemia with myelodysplasia-related changes diagnosed with multilineage dysplasia alone demonstrates a superior clinical outcome.Hum Pathol. 2020 Oct;104:117-126. doi: 10.1016/j.humpath.2020.08.003. Epub 2020 Aug 13. Hum Pathol. 2020. PMID: 32798550
-
Characteristics of acute myeloid leukemia with myelodysplasia-related changes: A retrospective analysis in a cohort of Chinese patients.Am J Hematol. 2014 Sep;89(9):874-81. doi: 10.1002/ajh.23772. Epub 2014 Jun 19. Am J Hematol. 2014. PMID: 24861848
-
Myelodysplastic syndrome in children: differentiation from acute myeloid leukemia with a low blast count.Leukemia. 1997 Feb;11(2):206-11. doi: 10.1038/sj.leu.2400558. Leukemia. 1997. PMID: 9009082 Review.
-
Acute Myeloid Leukemia With Myelodysplasia-Related Changes.Am J Clin Pathol. 2015 Jul;144(1):29-43. doi: 10.1309/AJCP58RSMFRHLHHH. Am J Clin Pathol. 2015. PMID: 26071460 Review.
Cited by
-
An Unusually Short Latent Period of Therapy-Related Myeloid Neoplasm Harboring a Rare MLL-EP300 Rearrangement: Case Report and Literature Review.Case Rep Hematol. 2019 Oct 2;2019:4532434. doi: 10.1155/2019/4532434. eCollection 2019. Case Rep Hematol. 2019. PMID: 31662917 Free PMC article.
-
The latest edition of WHO and ELN guidance and a new risk model for Chinese acute myeloid leukemia patients.Front Med (Lausanne). 2023 Jun 23;10:1165445. doi: 10.3389/fmed.2023.1165445. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37435533 Free PMC article.
-
Secondary AML Emerging After Therapy with Hypomethylating Agents: Outcomes, Prognostic Factors, and Treatment Options.Curr Hematol Malig Rep. 2021 Feb;16(1):97-111. doi: 10.1007/s11899-021-00608-6. Epub 2021 Feb 20. Curr Hematol Malig Rep. 2021. PMID: 33609248 Review.
-
Emerging Therapies for Acute Myelogenus Leukemia Patients Targeting Apoptosis and Mitochondrial Metabolism.Cancers (Basel). 2019 Feb 22;11(2):260. doi: 10.3390/cancers11020260. Cancers (Basel). 2019. PMID: 30813354 Free PMC article. Review.
-
Allogeneic hematopoietic cell transplantation can overcome the adverse prognosis indicated by secondary-type mutations in de novo acute myeloid leukemia.Bone Marrow Transplant. 2022 Dec;57(12):1810-1819. doi: 10.1038/s41409-022-01817-0. Epub 2022 Sep 23. Bone Marrow Transplant. 2022. PMID: 36151367
References
-
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105-117. - PubMed
-
- Weinberg OK, Seetharam M, Ren L, et al. . Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood. 2009;113(9):1906-1908. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

