Does Size Matter? Comparison of Extraction Yields for Different-Sized DNA Fragments by Seven Different Routine and Four New Circulating Cell-Free Extraction Methods
- PMID: 30282788
- PMCID: PMC6258844
- DOI: 10.1128/JCM.01061-18
Does Size Matter? Comparison of Extraction Yields for Different-Sized DNA Fragments by Seven Different Routine and Four New Circulating Cell-Free Extraction Methods
Abstract
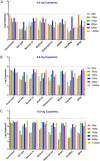
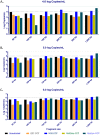
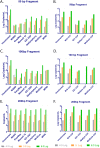
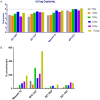
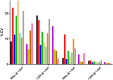
An element essential for PCR detection of microbial agents in many sample types is the extraction step, designed to purify nucleic acids. Despite the importance of this step, yields have not been extensively compared across methods to determine whether the method used contributes to quantitative differences and the lack of commutability seen with existing clinical methods. This may in part explain why plasma and blood viral load assays have proven difficult to standardize. Also, studies have identified small DNA fragments of <200 bp in plasma (cell-free DNA [cfDNA]), which may include significant quantities of viral DNA. Our study evaluated extraction yields for 11 commercially available extraction methods, including 4 new methods designed to isolate cfDNA. Solutions of DNA fragments with sizes ranging from 50 to 1,500 bp were extracted, and then the eluates were tested by droplet digital PCR to determine the DNA fragment yield for each method. The results demonstrated a wide range of extraction yields across the variety of methods/instruments used, with the 50- and 100-bp fragment sizes showing especially inconsistent quantitative results and poor yields of less than 20%. Slightly higher, more consistent yields were seen with 2 of the 4 circulating cell-free extraction kits. These results demonstrate a significant need for further evaluation of nucleic acid yields across the variety of extraction platforms and highlight the poor extraction yields of small DNA fragments by existing methods. Further work is necessary to determine the impact of this inconsistency across instruments and the relevance of the low yields for smaller DNA fragments in clinical virology testing.
Keywords: ccfDNA; extraction methods; extraction yield; viral DNA processing; viral diagnostics.
Copyright © 2018 American Society for Microbiology.
Figures
References
-
- Cornelissen M, Gall A, Vink M, Zorgdrager F, Binter S, Edwards S, Jurriaans S, Bakker M, Ong SH, Gras L, van Sighem A, Bezemer D, de Wolf F, Reiss P, Kellam P, Berkhout B, Fraser C, van der Kuyl AC, Consortium B. 2017. From clinical sample to complete genome: comparing methods for the extraction of HIV-1 RNA for high-throughput deep sequencing. Virus Res 239:10–16. doi:10.1016/j.virusres.2016.08.004. - DOI - PubMed
-
- Stevens W, Horsfield P, Scott LE. 2007. Evaluation of the performance of the automated NucliSENS easyMAG and EasyQ systems versus the Roche AmpliPrep-AMPLICOR combination for high-throughput monitoring of human immunodeficiency virus load. J Clin Microbiol 45:1244–1249. doi:10.1128/JCM.01540-06. - DOI - PMC - PubMed
-
- Swanson P, Holzmayer V, Huang S, Hay P, Adebiyi A, Rice P, Abravaya K, Thamm S, Devare SG, Hackett J Jr. 2006. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from London: comparison to VERSANT HIV-1 RNA 3.0, AMPLICOR HIV-1 MONITOR v1.5, and LCx HIV RNA quantitative assays. J Virol Methods 137:184–192. doi:10.1016/j.jviromet.2006.06.010. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

