OTUB1 non-catalytically stabilizes the E2 ubiquitin-conjugating enzyme UBE2E1 by preventing its autoubiquitination
- PMID: 30282802
- PMCID: PMC6254341
- DOI: 10.1074/jbc.RA118.004677
OTUB1 non-catalytically stabilizes the E2 ubiquitin-conjugating enzyme UBE2E1 by preventing its autoubiquitination
Abstract
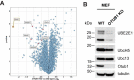
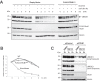
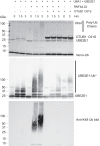
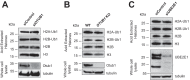
OTUB1 is a deubiquitinating enzyme that cleaves Lys-48-linked polyubiquitin chains and also regulates ubiquitin signaling through a unique, noncatalytic mechanism. OTUB1 binds to a subset of E2 ubiquitin-conjugating enzymes and inhibits their activity by trapping the E2∼ubiquitin thioester and preventing ubiquitin transfer. The same set of E2s stimulate the deubiquitinating activity of OTUB1 when the E2 is not charged with ubiquitin. Previous studies have shown that, in cells, OTUB1 binds to E2-conjugating enzymes of the UBE2D (UBCH5) and UBE2E families, as well as to UBE2N (UBC13). Cellular roles have been identified for the interaction of OTUB1 with UBE2N and members of the UBE2D family, but not for interactions with UBE2E E2 enzymes. We report here a novel role for OTUB1-E2 interactions in modulating E2 protein ubiquitination. We observe that Otub1-/- knockout mice exhibit late-stage embryonic lethality. We find that OTUB1 depletion dramatically destabilizes the E2-conjugating enzyme UBE2E1 (UBCH6) in both mouse and human OTUB1 knockout cell lines. Of note, this effect is independent of the catalytic activity of OTUB1, but depends on its ability to bind to UBE2E1. We show that OTUB1 suppresses UBE2E1 autoubiquitination in vitro and in cells, thereby preventing UBE2E1 from being targeted to the proteasome for degradation. Taken together, we provide evidence that OTUB1 rescues UBE2E1 from degradation in vivo.
Keywords: UBE2E1; deubiquitylation (deubiquitination); histone; ubiquitin; ubiquitin thioesterase (OTUB1); ubiquitin-conjugating enzyme (E2 enzyme).
© 2018 Pasupala et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

