A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5α and mammalian autophagy-related ATG8 proteins
- PMID: 30282803
- PMCID: PMC6254359
- DOI: 10.1074/jbc.RA118.004202
A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5α and mammalian autophagy-related ATG8 proteins
Abstract
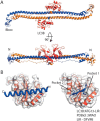
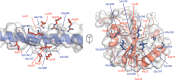
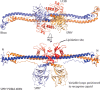
The retroviral restriction factor tripartite motif-containing 5α (Trim5α) acts during the early postentry stages of the retroviral life cycle to block infection by a broad range of retroviruses, disrupting reverse transcription and integration. The mechanism of this restriction is poorly understood, but it has recently been suggested to involve recruitment of components of the autophagy machinery, including members of the mammalian autophagy-related 8 (ATG8) family involved in targeting proteins to the autophagosome. To better understand the molecular details of this interaction, here we utilized analytical ultracentrifugation to characterize the binding of six ATG8 isoforms and determined the crystal structure of the Trim5α Bbox coiled-coil region in complex with one member of the mammalian ATG8 proteins, autophagy-related protein LC3 B (LC3B). We found that Trim5α binds all mammalian ATG8s and that, unlike the typical LC3-interacting region (LIR) that binds to mammalian ATG8s through a β-strand motif comprising approximately six residues, LC3B binds to Trim5α via the α-helical coiled-coil region. The orientation of the structure demonstrated that LC3B could be accommodated within a Trim5α assembly that can bind the retroviral capsid. However, mutation of the binding interface does not affect retroviral restriction. Comparison of the typical linear β-strand LIR with our atypical helical LIR reveals a conservation of the presentation of residues that are required for the interaction with LC3B. This observation expands the range of LC3B-binding proteins to include helical binding motifs and demonstrates a link between Trim5α and components of the autophagosome.
Keywords: LC3B; LIR; Trim5alpha; antiviral protein; autophagy; human immunodeficiency virus (HIV); protein complex; protein-protein interaction; retroviral restriction factor; retrovirus.
© 2018 Keown et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Birgisdottir Å. B., Lamark T., and Johansen T. (2013) The LIR motif: crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 - PubMed
-
- McEwan D. G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F. P., Miranda de Stegmann D., Bhogaraju S., Maddi K., Gatti E., Helfrich M. H., Wakatsuki S., Behrends C., Pierre P., et al. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54 10.1016/j.molcel.2014.11.006 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

