Aneuploidy in intestinal stem cells promotes gut dysplasia in Drosophila
- PMID: 30282810
- PMCID: PMC6219720
- DOI: 10.1083/jcb.201804205
Aneuploidy in intestinal stem cells promotes gut dysplasia in Drosophila
Abstract
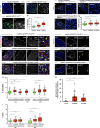
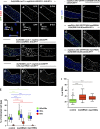
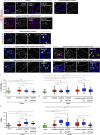
Aneuploidy is associated with different human diseases including cancer. However, different cell types appear to respond differently to aneuploidy, either by promoting tumorigenesis or causing cell death. We set out to study the behavior of adult Drosophila melanogaster intestinal stem cells (ISCs) after induction of chromosome missegregation either by abrogation of the spindle assembly checkpoint or through kinetochore disruption or centrosome amplification. These conditions induce moderate levels of aneuploidy in ISCs, and we find no evidence of apoptosis. Instead, we observe a significant accumulation of ISCs associated with increased stem cell proliferation and an excess of enteroendocrine cells. Moreover, aneuploidy causes up-regulation of the JNK pathway throughout the posterior midgut, and specific inhibition of JNK signaling in ISCs is sufficient to prevent dysplasia. Our findings highlight the importance of understanding the behavior of different stem cell populations to aneuploidy and how these can act as reservoirs for genomic alterations that can lead to tissue pathologies.
© 2018 Resende et al.
Figures





Similar articles
-
Aneuploidy facilitates dysplastic and tumorigenic phenotypes in the Drosophila gut.Biol Open. 2021 Nov 15;10(11):bio058623. doi: 10.1242/bio.058623. Epub 2021 Nov 3. Biol Open. 2021. PMID: 33948620 Free PMC article.
-
Metformin inhibits age-related centrosome amplification in Drosophila midgut stem cells through AKT/TOR pathway.Mech Ageing Dev. 2015 Jul;149:8-18. doi: 10.1016/j.mad.2015.05.004. Epub 2015 May 16. Mech Ageing Dev. 2015. PMID: 25988874
-
The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila.Aging (Albany NY). 2009 May 21;1(7):637-51. doi: 10.18632/aging.100054. Aging (Albany NY). 2009. PMID: 20157545 Free PMC article.
-
Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer.Adv Exp Med Biol. 2005;570:393-421. doi: 10.1007/1-4020-3764-3_14. Adv Exp Med Biol. 2005. PMID: 18727509 Review.
-
Centrosomes, chromosome instability (CIN) and aneuploidy.Curr Opin Cell Biol. 2012 Dec;24(6):809-15. doi: 10.1016/j.ceb.2012.10.006. Epub 2012 Nov 3. Curr Opin Cell Biol. 2012. PMID: 23127609 Free PMC article. Review.
Cited by
-
Understanding How Genetic Mutations Collaborate with Genomic Instability in Cancer.Cells. 2021 Feb 6;10(2):342. doi: 10.3390/cells10020342. Cells. 2021. PMID: 33562057 Free PMC article. Review.
-
Simple aneuploidy evades p53 surveillance and promotes niche factor-independent growth in human intestinal organoids.Mol Biol Cell. 2024 Aug 1;35(8):br15. doi: 10.1091/mbc.E24-04-0166. Epub 2024 Jul 10. Mol Biol Cell. 2024. PMID: 38985518 Free PMC article.
-
PLK4: a promising target for cancer therapy.J Cancer Res Clin Oncol. 2019 Oct;145(10):2413-2422. doi: 10.1007/s00432-019-02994-0. Epub 2019 Sep 6. J Cancer Res Clin Oncol. 2019. PMID: 31492983 Free PMC article. Review.
-
Cohesin controls intestinal stem cell identity by maintaining association of Escargot with target promoters.Elife. 2020 Feb 5;9:e48160. doi: 10.7554/eLife.48160. Elife. 2020. PMID: 32022682 Free PMC article.
-
Induced aneuploidy in neural stem cells triggers a delayed stress response and impairs adult life span in flies.PLoS Biol. 2019 Feb 22;17(2):e3000016. doi: 10.1371/journal.pbio.3000016. eCollection 2019 Feb. PLoS Biol. 2019. PMID: 30794535 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

