Metabolomic Analyses Reveal Extensive Progenitor Cell Deficiencies in a Mouse Model of Duchenne Muscular Dystrophy
- PMID: 30282911
- PMCID: PMC6315702
- DOI: 10.3390/metabo8040061
Metabolomic Analyses Reveal Extensive Progenitor Cell Deficiencies in a Mouse Model of Duchenne Muscular Dystrophy
Abstract
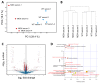
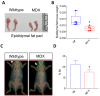
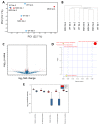
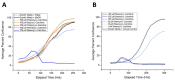
Duchenne muscular dystrophy (DMD) is a musculoskeletal disorder that causes severe morbidity and reduced lifespan. Individuals with DMD have an X-linked mutation that impairs their ability to produce functional dystrophin protein in muscle. No cure exists for this disease and the few therapies that are available do not dramatically delay disease progression. Thus, there is a need to better understand the mechanisms underlying DMD which may ultimately lead to improved treatment options. The muscular dystrophy (MDX) mouse model is frequently used to explore DMD disease traits. Though some studies of metabolism in dystrophic mice exist, few have characterized metabolic profiles of supporting cells in the diseased environment. Using nontargeted metabolomics we characterized metabolic alterations in muscle satellite cells (SCs) and serum of MDX mice. Additionally, live-cell imaging revealed MDX-derived adipose progenitor cell (APC) defects. Finally, metabolomic studies revealed a striking elevation of acylcarnitines in MDX APCs, which we show can inhibit APC proliferation. Together, these studies highlight widespread metabolic alterations in multiple progenitor cell types and serum from MDX mice and implicate dystrophy-associated metabolite imbalances in APCs as a potential contributor to adipose tissue disequilibrium in DMD.
Keywords: Duchenne muscular dystrophy; adipose tissue; metabolomics; skeletal muscle; stem cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy.Exp Physiol. 2014 Apr;99(4):675-87. doi: 10.1113/expphysiol.2013.077255. Epub 2014 Jan 17. Exp Physiol. 2014. PMID: 24443351
-
Differential metabolic secretion between muscular dystrophy mouse-derived spindle cell sarcomas and rhabdomyosarcomas drives tumor type development.Am J Physiol Cell Physiol. 2024 Jul 1;327(1):C34-C47. doi: 10.1152/ajpcell.00523.2023. Epub 2024 Apr 22. Am J Physiol Cell Physiol. 2024. PMID: 38646787
-
Pre-clinical evaluation of N-acetylcysteine reveals side effects in the mdx mouse model of Duchenne muscular dystrophy.J Physiol. 2017 Dec 1;595(23):7093-7107. doi: 10.1113/JP274229. Epub 2017 Sep 30. J Physiol. 2017. PMID: 28887840 Free PMC article.
-
Current Translational Research and Murine Models For Duchenne Muscular Dystrophy.J Neuromuscul Dis. 2016 Mar 3;3(1):29-48. doi: 10.3233/JND-150113. J Neuromuscul Dis. 2016. PMID: 27854202 Free PMC article. Review.
-
Brain function in Duchenne muscular dystrophy.Brain. 2002 Jan;125(Pt 1):4-13. doi: 10.1093/brain/awf012. Brain. 2002. PMID: 11834588 Review.
Cited by
-
Caveolin and NOS in the Development of Muscular Dystrophy.Int J Mol Sci. 2024 Aug 12;25(16):8771. doi: 10.3390/ijms25168771. Int J Mol Sci. 2024. PMID: 39201459 Free PMC article. Review.
-
Muscle stem cells contribute to long-term tissue repletion following surgical sepsis.J Cachexia Sarcopenia Muscle. 2023 Jun;14(3):1424-1440. doi: 10.1002/jcsm.13214. Epub 2023 Mar 8. J Cachexia Sarcopenia Muscle. 2023. PMID: 36883680 Free PMC article.
-
A comparison of the bone and growth phenotype of mdx, mdx:Cmah-/- and mdx:Utrn+/- murine models with the C57BL/10 wild-type mouse.Dis Model Mech. 2020 Jan 10;13(2):dmm040659. doi: 10.1242/dmm.040659. Dis Model Mech. 2020. PMID: 31754018 Free PMC article.
-
Single-cell deconstruction of post-sepsis skeletal muscle and adipose tissue microenvironments.J Cachexia Sarcopenia Muscle. 2020 Oct;11(5):1351-1363. doi: 10.1002/jcsm.12596. Epub 2020 Jul 8. J Cachexia Sarcopenia Muscle. 2020. PMID: 32643301 Free PMC article.
-
The metabolomic plasma profile of patients with Duchenne muscular dystrophy: providing new evidence for its pathogenesis.Orphanet J Rare Dis. 2023 Sep 5;18(1):273. doi: 10.1186/s13023-023-02885-1. Orphanet J Rare Dis. 2023. PMID: 37670327 Free PMC article.
References
-
- Leikina E.S., Plotnikov N.N., Prokopenko L.I. Basic methods of prevention of helminthiases, their improvement and development. Med. Parazitol. (Mosk) 1974;43:259–265. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

