Protein Quality Control in the Endoplasmic Reticulum and Cancer
- PMID: 30282948
- PMCID: PMC6213883
- DOI: 10.3390/ijms19103020
Protein Quality Control in the Endoplasmic Reticulum and Cancer
Abstract
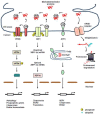
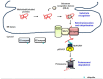
The endoplasmic reticulum (ER) is an essential compartment of the biosynthesis, folding, assembly, and trafficking of secretory and transmembrane proteins, and consequently, eukaryotic cells possess specialized machineries to ensure that the ER enables the proteins to acquire adequate folding and maturation for maintaining protein homeostasis, a process which is termed proteostasis. However, a large variety of physiological and pathological perturbations lead to the accumulation of misfolded proteins in the ER, which is referred to as ER stress. To resolve ER stress and restore proteostasis, cells have evolutionary conserved protein quality-control machineries of the ER, consisting of the unfolded protein response (UPR) of the ER, ER-associated degradation (ERAD), and autophagy. Furthermore, protein quality-control machineries of the ER play pivotal roles in the control of differentiation, progression of cell cycle, inflammation, immunity, and aging. Therefore, severe and non-resolvable ER stress is closely associated with tumor development, aggressiveness, and response to therapies for cancer. In this review, we highlight current knowledge in the molecular understanding and physiological relevance of protein quality control of the ER and discuss new insights into how protein quality control of the ER is implicated in the pathogenesis of cancer, which could contribute to therapeutic intervention in cancer.
Keywords: ER-associated protein degradation (ERAD), autophagy; cancer; endoplasmic reticulum (ER) stress; protein quality control; proteostasis; unfolded protein response (UPR) of the ER.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

