LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212
- PMID: 30282972
- PMCID: PMC6170498
- DOI: 10.1038/s41419-018-1068-x
LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212
Erratum in
-
Correction: LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212.Cell Death Dis. 2022 Feb 9;13(2):133. doi: 10.1038/s41419-022-04558-6. Cell Death Dis. 2022. PMID: 35140199 Free PMC article. No abstract available.
Abstract
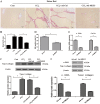
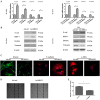
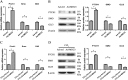
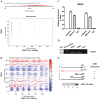
Activation of hepatic stellate cells (HSCs), a pivotal event in liver fibrosis, is considered as an epithelial-mesenchymal transition (EMT) process. Deregulation of long noncoding RNAs (lncRNAs) has been reported to be involved in a series of human diseases. LncRNA-maternally expressed gene 3 (MEG3) functions as a tumor suppressor in cancers and has been shown to play a vital role in EMT process. However, the biological role of MEG3 in liver fibrosis is largely unknown. In this study, MEG3 was reduced in vivo and in vitro during liver fibrosis. Restoring of MEG3 expression led to the suppression of liver fibrosis, with a reduction in α-SMA and type I collagen. Notably, MEG3 overexpression inhibited HSC activation through EMT, associated with an increase in epithelial markers and a reduction in mesenchymal markers. Further studies showed that Hedgehog (Hh) pathway-mediated EMT process was involved in the effects of MEG3 on HSC activation. Smoothened (SMO) is a member of Hh pathway. Using bioinformatic analysis, an interaction between MEG3 and SMO protein was predicted. This interaction was confirmed by the results of RNA immunoprecipitation and deletion-mapping analysis. Furthermore, MEG3 was confirmed as a target of microRNA-212 (miR-212). miR-212 was partly responsible for the effects of MEG3 on EMT process. Interestingly, MEG3 was also reduced in chronic hepatitis B (CHB) patients with liver fibrosis when compared with healthy controls. MEG3 negatively correlated with fibrosis stage in CHB patients. In conclusion, we demonstrate that MEG3 inhibits Hh-mediated EMT process in liver fibrosis via SMO protein and miR-212.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

