Inhibition of the gyrA promoter by transcription-coupled DNA supercoiling in Escherichia coli
- PMID: 30282997
- PMCID: PMC6170449
- DOI: 10.1038/s41598-018-33089-4
Inhibition of the gyrA promoter by transcription-coupled DNA supercoiling in Escherichia coli
Abstract
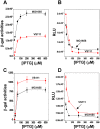
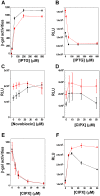
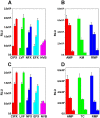
The E. coli gyrA promoter (PgyrA) is a DNA supercoiling sensitive promoter, stimulated by relaxation of DNA templates, and inhibited by (-) DNA supercoiling in bacteria. However, whether PgyrA can be inhibited by transient and localized transcription-coupled DNA supercoiling (TCDS) has not been fully examined. In this paper, using different DNA templates including the E. coli chromosome, we show that transient and localized TCDS strongly inhibits PgyrA in E. coli. This result can be explained by a twin-supercoiled domain model of transcription in which (+) and (-) supercoiled domains are generated around the transcribing RNA polymerase. We also find that fluoroquinolones, such as ciprofloxacin, can substantially increase the expression of the firefly luciferase under the control of the PgyrA coupled to a divergent IPTG-inducible promoter in the presence of IPTG. This stimulation of PgyrA by fluoroquinolones can be also explained by the twin-supercoiled domain model of transcription. This unique property of TCDS may be configured into a high throughput-screening (HTS) assay to identify antimicrobial compounds targeting bacterial DNA gyrase.
Conflict of interest statement
A US patent has been awarded to authors related to this manuscript. Patent title: Materials and Methods for identifying gyrase inhibitors. US Patent Number: 10000807.
Figures


Similar articles
-
Transient and dynamic DNA supercoiling potently stimulates the leu-500 promoter in Escherichia coli.J Biol Chem. 2017 Sep 1;292(35):14566-14575. doi: 10.1074/jbc.M117.794628. Epub 2017 Jul 10. J Biol Chem. 2017. PMID: 28696257 Free PMC article.
-
Dependence of transcription-coupled DNA supercoiling on promoter strength in Escherichia coli topoisomerase I deficient strains.Gene. 2013 Feb 10;514(2):82-90. doi: 10.1016/j.gene.2012.11.011. Epub 2012 Nov 29. Gene. 2013. PMID: 23201416 Free PMC article.
-
Discoordinate gene expression of gyrA and gyrB in response to DNA gyrase inhibition in Escherichia coli.J Basic Microbiol. 1997;37(1):53-69. doi: 10.1002/jobm.3620370109. J Basic Microbiol. 1997. PMID: 9090126
-
[DNA supercoiling and topoisomerases in Escherichia coli].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):291-304. Rev Latinoam Microbiol. 1995. PMID: 8850348 Review. Spanish.
-
Local DNA topology and gene expression: the case of the leu-500 promoter.Mol Microbiol. 1991 Apr;5(4):779-83. doi: 10.1111/j.1365-2958.1991.tb00749.x. Mol Microbiol. 1991. PMID: 1857204 Review.
Cited by
-
Psoralen mapping reveals a bacterial genome supercoiling landscape dominated by transcription.Nucleic Acids Res. 2022 May 6;50(8):4436-4449. doi: 10.1093/nar/gkac244. Nucleic Acids Res. 2022. PMID: 35420137 Free PMC article.
-
Transcriptional Responses of Pseudomonas aeruginosa to Inhibition of Lipoprotein Transport by a Small Molecule Inhibitor.J Bacteriol. 2020 Nov 19;202(24):e00452-20. doi: 10.1128/JB.00452-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32989085 Free PMC article.
-
DNA Supercoiling: an Ancestral Regulator of Gene Expression in Pathogenic Bacteria?Comput Struct Biotechnol J. 2019 Jul 26;17:1047-1055. doi: 10.1016/j.csbj.2019.07.013. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31452857 Free PMC article. Review.
-
DNA supercoiling and transcription in bacteria: a two-way street.BMC Mol Cell Biol. 2019 Jul 18;20(1):26. doi: 10.1186/s12860-019-0211-6. BMC Mol Cell Biol. 2019. PMID: 31319794 Free PMC article. Review.
-
MoCloFlex: A Modular Yet Flexible Cloning System.Front Bioeng Biotechnol. 2019 Oct 17;7:271. doi: 10.3389/fbioe.2019.00271. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31750294 Free PMC article.
References
-
- Bates, A. D. & Maxwell, A. DNA Topology (Oxford University Press, Oxford, UK, 2005).
-
- James, C. Wang Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases (Cold Spring Harbor Laboratory Press, 2008).
-
- Cozzarelli, N. R. & Wang, J. C. DNA Topology and Its Biological Effects (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1990).
-
- Snoep JL, van der Weijden CC, Andersen HW, Westerhoff HV, Jensen PR. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 2002;269:1662–1669. doi: 10.1046/j.1432-1327.2002.02803.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

