OTX2 restricts entry to the mouse germline
- PMID: 30283136
- PMCID: PMC6485399
- DOI: 10.1038/s41586-018-0581-5
OTX2 restricts entry to the mouse germline
Abstract
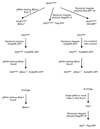
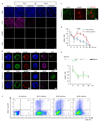
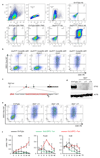
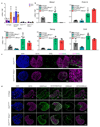
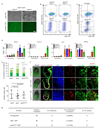
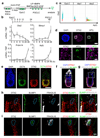
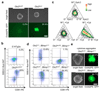
The successful segregation of germ cells from somatic lineages is vital for sexual reproduction and species survival. In the mouse, primordial germ cells (PGCs), precursors of all germ cells, are induced from the post-implantation epiblast1. Induction requires BMP4 signalling to prospective PGCs2 and the intrinsic action of PGC transcription factors3-6. However, the molecular mechanisms that connect BMP4 to induction of the PGC transcription factors that are responsible for segregating PGCs from somatic lineages are unknown. Here we show that the transcription factor OTX2 is a key regulator of these processes. Downregulation of Otx2 precedes the initiation of the PGC programme both in vitro and in vivo. Deletion of Otx2 in vitro markedly increases the efficiency of PGC-like cell differentiation and prolongs the period of PGC competence. In the absence of Otx2 activity, differentiation of PGC-like cells becomes independent of the otherwise essential cytokine signals, with germline entry initiating even in the absence of the PGC transcription factor BLIMP1. Deletion of Otx2 in vivo increases PGC numbers. These data demonstrate that OTX2 functions repressively upstream of PGC transcription factors, acting as a roadblock to limit entry of epiblast cells to the germline to a small window in space and time, thereby ensuring correct numerical segregation of germline cells from the soma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Comment in
-
How to lose your inheritance.Nature. 2018 Oct;562(7728):497-498. doi: 10.1038/d41586-018-06849-5. Nature. 2018. PMID: 30349122 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

