Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD
- PMID: 30283140
- PMCID: PMC6309448
- DOI: 10.1038/s41586-018-0587-z
Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD
Abstract
Pathogenic bacteria such as Escherichia coli assemble surface structures termed pili, or fimbriae, to mediate binding to host-cell receptors1. Type 1 pili are assembled via the conserved chaperone-usher pathway2-5. The outer-membrane usher FimD recruits pilus subunits bound by the chaperone FimC via the periplasmic N-terminal domain of the usher. Subunit translocation through the β-barrel channel of the usher occurs at the two C-terminal domains (which we label CTD1 and CTD2) of this protein. How the chaperone-subunit complex bound to the N-terminal domain is handed over to the C-terminal domains, as well as the timing of subunit polymerization into the growing pilus, have previously been unclear. Here we use cryo-electron microscopy to capture a pilus assembly intermediate (FimD-FimC-FimF-FimG-FimH) in a conformation in which FimD is in the process of handing over the chaperone-bound end of the growing pilus to the C-terminal domains. In this structure, FimF has already polymerized with FimG, and the N-terminal domain of FimD swings over to bind CTD2; the N-terminal domain maintains contact with FimC-FimF, while at the same time permitting access to the C-terminal domains. FimD has an intrinsically disordered N-terminal tail that precedes the N-terminal domain. This N-terminal tail folds into a helical motif upon recruiting the FimC-subunit complex, but reorganizes into a loop to bind CTD2 during handover. Because both the N-terminal and C-terminal domains of FimD are bound to the end of the growing pilus, the structure further suggests a mechanism for stabilizing the assembly intermediate to prevent the pilus fibre diffusing away during the incorporation of thousands of subunits.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing financial interests.
Figures






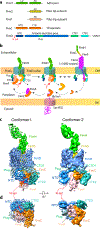


Similar articles
-
Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD.FEBS Lett. 2008 Mar 5;582(5):651-5. doi: 10.1016/j.febslet.2008.01.030. Epub 2008 Jan 31. FEBS Lett. 2008. PMID: 18242189
-
Structures of the Escherichia coli type 1 pilus during pilus rod assembly and after assembly termination.Nat Commun. 2025 May 29;16(1):4988. doi: 10.1038/s41467-025-60325-z. Nat Commun. 2025. PMID: 40442073 Free PMC article.
-
Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD.J Mol Biol. 2003 Jul 11;330(3):513-25. doi: 10.1016/s0022-2836(03)00591-6. J Mol Biol. 2003. PMID: 12842468
-
Crystallography and electron microscopy of chaperone/usher pilus systems.Adv Exp Med Biol. 2011;715:159-74. doi: 10.1007/978-94-007-0940-9_10. Adv Exp Med Biol. 2011. PMID: 21557063 Review.
-
Chaperone-assisted assembly and molecular architecture of adhesive pili.Annu Rev Microbiol. 1991;45:383-415. doi: 10.1146/annurev.mi.45.100191.002123. Annu Rev Microbiol. 1991. PMID: 1683764 Review.
Cited by
-
Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.Nature. 2022 Sep;609(7926):335-340. doi: 10.1038/s41586-022-05095-0. Epub 2022 Jul 19. Nature. 2022. PMID: 35853476 Free PMC article.
-
Catheter-Associated Urinary Tract Infections: Current Challenges and Future Prospects.Res Rep Urol. 2022 Apr 4;14:109-133. doi: 10.2147/RRU.S273663. eCollection 2022. Res Rep Urol. 2022. PMID: 35402319 Free PMC article. Review.
-
Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.Nat Commun. 2021 Sep 1;12(1):5207. doi: 10.1038/s41467-021-25522-6. Nat Commun. 2021. PMID: 34471127 Free PMC article.
-
Conformational ensembles in Klebsiella pneumoniae FimH impact uropathogenesis.Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2409655121. doi: 10.1073/pnas.2409655121. Epub 2024 Sep 17. Proc Natl Acad Sci U S A. 2024. PMID: 39288182 Free PMC article.
-
Stochastic chain termination in bacterial pilus assembly.Nat Commun. 2023 Nov 24;14(1):7718. doi: 10.1038/s41467-023-43449-y. Nat Commun. 2023. PMID: 38001074 Free PMC article.
References
-
- Thanassi DG, Saulino ET & Hultgren SJ The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol 1, 223–231 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

