Parallel phylogeography of Prochlorococcus and Synechococcus
- PMID: 30283146
- PMCID: PMC6331572
- DOI: 10.1038/s41396-018-0287-6
Parallel phylogeography of Prochlorococcus and Synechococcus
Abstract
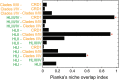
The globally abundant marine Cyanobacteria Prochlorococcus and Synechococcus share many physiological traits but presumably have different evolutionary histories and associated phylogeography. In Prochlorococcus, there is a clear phylogenetic hierarchy of ecotypes, whereas multiple Synechococcus clades have overlapping physiologies and environmental distributions. However, microbial traits are associated with different phylogenetic depths. Using this principle, we reclassified diversity at different phylogenetic levels and compared the phylogeography. We sequenced the genetic diversity of Prochlorococcus and Synechococcus from 339 samples across the tropical Pacific Ocean and North Atlantic Ocean using a highly variable phylogenetic marker gene (rpoC1). We observed clear parallel niche distributions of ecotypes leading to high Pianka's Index values driven by distinct shifts at two transition points. The first transition point at 6°N distinguished ecotypes adapted to warm waters but separated by macronutrient content. At 39°N, ecotypes adapted to warm, low macronutrient vs. colder, high macronutrient waters shifted. Finally, we detected parallel vertical and regional single-nucleotide polymorphism microdiversity within clades from both Prochlorococcus and Synechococcus, suggesting uniquely adapted populations at very specific depths, as well as between the Atlantic and Pacific Oceans. Overall, this study demonstrates that Prochlorococcus and Synechococcus have shared phylogenetic organization of traits and associated phylogeography.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52:2205–20. doi: 10.4319/lo.2007.52.5.2205. - DOI

