Molecular Background of Pi Deficiency-Induced Root Hair Growth in Brassica carinata - A Fasciclin-Like Arabinogalactan Protein Is Involved
- PMID: 30283481
- PMCID: PMC6157447
- DOI: 10.3389/fpls.2018.01372
Molecular Background of Pi Deficiency-Induced Root Hair Growth in Brassica carinata - A Fasciclin-Like Arabinogalactan Protein Is Involved
Abstract
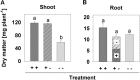
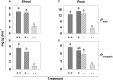
Formation of longer root hairs under limiting phosphate (P) conditions can increase the inorganic P (Pi) uptake. Here, regulatory candidate genes for Pi deficiency-induced root hair growth were identified by comparison of massive analysis of cDNA ends (MACE) provided expression profiles of two Brassica carinata cultivars (cv.) differing in their root hair response to Pi deficiency: cv. Bale develops longer root hairs under Pi deficiency, but not cv. Bacho. A split-root experiment was conducted for the differentiation between locally and systemically regulated genes. Furthermore, plants were exposed to nitrogen and potassium deficiency to identify P-specific reacting genes. The latter were knocked out by CRISPR/Cas9 and the effect on the root hair length was determined. About 500 genes were differentially expressed under Pi deficiency in cv. Bale, while these genes did not respond to the low P supply in cv. Bacho. Thirty-three candidate genes with a potential regulatory role were selected and the transcriptional regulation of 30 genes was confirmed by quantitative PCR. Only five candidate genes seemed to be either exclusively regulated locally (two) or systemically (three), whereas 25 genes seemed to be involved in both local and systemic signaling pathways. Potassium deficiency affected neither the root hair length nor the expression of the 30 candidate genes. By contrast, both P and nitrogen deficiency increased the root hair length, and both affected the transcript levels in 26 cases. However, four genes reacted specifically to Pi starvation. These genes and, additionally, INORGANIC PHOSPHATE TRANSPORTER 1 (BcPHT1) were targeted by CRISPR/Cas9. However, even if the transcript levels of five of these genes were clearly decreased, FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 1 (BcFLA1) was the only gene whose downregulation reduced the root hair length in transgenic hairy roots under Pi-deficient conditions. To the best of our knowledge, this is the first study describing a fasciclin-like arabinogalactan protein with a predicted role in the Pi deficiency-induced root hair elongation.
Keywords: BcFLA1; CRISPR/Cas9; K deficiency; MACE; N deficiency; hairy root; local and systemic response; split-root.
Figures












References
-
- Basu D., Tian L., Debrosse T., Poirier E., Emch K., Herock H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-Protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. 10.1371/journal.pone.0145092 - DOI - PMC - PubMed
-
- Bieleski R. L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 24 225–252.
-
- Bremer M. (2010). Molecular Mechanisms of Root Hair Growth Induced by Pi Deficiency in Brassica carinata. Doctoral thesis, Gottfried Wilhelm Leibniz Universität Hannover, Hannover.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

