Filter-Aided Sample Preparation Procedure for Mass Spectrometric Analysis of Plant Histones
- PMID: 30283482
- PMCID: PMC6156276
- DOI: 10.3389/fpls.2018.01373
Filter-Aided Sample Preparation Procedure for Mass Spectrometric Analysis of Plant Histones
Abstract
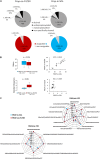
Characterization of histone post-translational modifications (PTMs) is still challenging, and robust histone sample preparation is essential for convincing evaluation of PTMs by mass spectrometry. An effective protocol for extracting plant histone proteins must also avoid excessive co-extraction of the numerous potential interfering compounds, including those related to secondary metabolism. Currently, the co-existence of histone marks is addressed mostly by shotgun proteomic analysis following chemical derivatization of histone lysine residues. Here, we report a straightforward approach for plant histone sample preparation for mass spectrometry, based on filter-aided sample preparation coupled with histone propionylation. The approach offers savings in sample handling and preparation time, enables removal of interfering compounds from the sample, and does not require either precipitation or dialysis of histone extract. We show the comparison of two protocol variants for derivatization of histone proteins, in-solution propionylation in the vial and propionylation on the filter unit. For both protocols, we obtained identical abundances of post-translationally modified histone peptides. Although shorter time is required for histone protein labeling on the filter unit, in-solution derivatization slightly outweighed filter-based variant by lower data variability. Nevertheless, both protocol variants appear to be efficient and convenient approach for preparation of plant histones for mass spectrometric analysis.
Keywords: Arabidopsis thaliana; epigenetics; filter-aided sample preparation; histone derivatization; mass spectrometry; post-translational modifications.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

