How the Conformations of an Internal Junction Contribute to Fold an RNA Domain
- PMID: 30285445
- PMCID: PMC6469688
- DOI: 10.1021/acs.jpcb.8b07262
How the Conformations of an Internal Junction Contribute to Fold an RNA Domain
Abstract
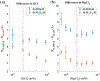
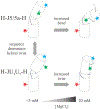
Like proteins, some RNAs fold to compact structures. We can model functional RNAs as a series of short, rigid, base-paired elements, connected by non-base-paired nucleotides that serve as junctions. These connecting regions bend and twist, facilitating the formation of tertiary contacts that stabilize compact states. Here, we explore the roles of salt and junction sequence in determining the structures of a ubiquitous connector: an asymmetric internal loop. We focus on the J5/5a junction from the widely studied P4-P6 domain of the Tetrahymena ribozyme. Following the addition of magnesium ions to fold P4-P6, this junction bends dramatically, bringing the two halves of the RNA domain together for tertiary contact engagement. Using single-molecule fluorescence resonance energy transfer (smFRET), we examine the role of sequence and salt on model RNA constructs that contain these junction regions. We explore the wild-type J5/5a junction as well as two sequence variants. These junctions display distinct, salt-dependent conformations. Small-angle X-ray scattering (SAXS) measurements verify that these effects persist in the full-length P4-P6 domain. These measurements underscore the importance of junction sequence and interactions with ions in facilitating RNA folding.
Figures
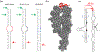

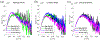

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

