Proteomic Investigation of Murine Neuronal α7-Nicotinic Acetylcholine Receptor Interacting Proteins
- PMID: 30285449
- PMCID: PMC6301012
- DOI: 10.1021/acs.jproteome.8b00618
Proteomic Investigation of Murine Neuronal α7-Nicotinic Acetylcholine Receptor Interacting Proteins
Abstract
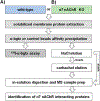
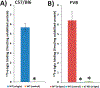
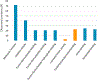
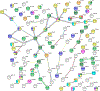
The α7-nicotinic acetylcholine receptor (α7-nAChR) is a ligand-gated ion channel that is expressed widely in vertebrates and is the principal high-affinity α-bungarotoxin (α-bgtx) binding protein in the mammalian CNS. α7-nAChRs associate with proteins that can modulate its properties. The α7-nAChR interactome is the summation of proteins interacting or associating with α7-nAChRs in a protein complex. To identify an α7-nAChR interactome in neural tissue, we isolated α-bgtx-affinity protein complexes from wild-type and α7-nAChR knockout (α7 KO) mouse whole brain tissue homogenates using α-bgtx-affinity beads. Affinity precipitated proteins were trypsinized and analyzed with an Orbitrap Fusion mass spectrometer. Proteins isolated with the α7-nAChR specific ligand, α-bgtx, were determined to be α7-nAChR associated proteins. The α7-nAChR subunit and 120 additional proteins were identified. Additionally, 369 proteins were identified as binding to α-bgtx in the absence of α7-nAChR expression, thereby identifying nonspecific proteins for α7-nAChR investigations using α-bgtx enrichment. These results expand on our previous investigations of α7-nAChR interacting proteins using α-bgtx-affinity bead isolation by controlling for differences between α7-nAChR and α-bgtx-specific proteins, developing an improved protein isolation methodology, and incorporating the latest technology in mass spectrometry. The α7-nAChR interactome identified in this study includes proteins associated with the expression, localization, function, or modulation of α7-nAChRs, and it provides a foundation for future studies to elucidate how these interactions contribute to human disease.
Keywords: Orbitrap Fusion; interactome; mass spectrometry; nAChR; nicotine; proteomics; α-bungarotoxin.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Dani JA; Bertrand D Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol 2007, 47 (1), 699–729. - PubMed
-
- Barry PH; Lynch JW Ligand-gated channels. IEEE transactions on nanobioscience 2005, 4 (1), 70–80. - PubMed
-
- Millar NS; Gotti C Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology 2009, 56 (1), 237–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

