Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments
- PMID: 30285603
- PMCID: PMC6167770
- DOI: 10.1186/s12864-018-5088-9
Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments
Abstract
Background: Pentatricopeptide-repeat proteins (PPRs) are characterized by tandem arrays of a degenerate 35-amino-acid (PPR motifs), which can bind RNA strands and participate in post-transcription. PPR proteins family is one of the largest families in land plants and play important roles in organelle RNA metabolism and plant development. However, the functions of PPR genes involved in biotic and abiotic stresses of rice (Oryza sativa L.) remain largely unknown.
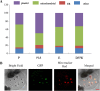
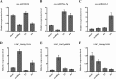
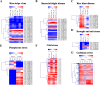
Results: In the present study, a comprehensive genome-wide analysis of PPR genes was performed. A total of 491 PPR genes were found in the rice genome, of which 246 PPR genes belong to the P subfamily, and 245 genes belong to the PLS subfamily. Gene structure analysis showed that most PPR genes lack intron. Chromosomal location analysis indicated that PPR genes were widely distributed in all 12 rice chromosomes. Phylogenetic relationship analysis revealed the distinct difference between the P and PLS subfamilies. Many PPR proteins are predicted to target chloroplasts or mitochondria, and a PPR protein (LOC_Os10g34310) was verified to localize in mitochondria. Furthermore, three PPR genes (LOC_Os03g17634,LOC_Os07g40820,LOC_Os04g51350) were verified as corresponding miRNA targets. The expression pattern analysis showed that many PPR genes could be induced under biotic and abiotic stresses. Finally, seven PPR genes were confirmed with their expression patterns under salinity or drought stress.
Conclusions: We found 491 PPR genes in the rice genome, and our genes structure analysis and syntenic analysis indicated that PPR genes might be derived from amplification by retro-transposition. The expression pattern present here suggested that PPR proteins have crucial roles in response to different abiotic stresses in rice. Taken together, our study provides a comprehensive analysis of the PPR gene family and will facilitate further studies on their roles in rice growth and development.
Keywords: Expression patterns; Pentatricopeptide-repeat protein; Rice (Oryza sativa L.); Salinity or drought stress; miRNA.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

