Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A
- PMID: 30285611
- PMCID: PMC6171223
- DOI: 10.1186/s12864-018-5113-z
Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A
Abstract
Background: MADS-box genes encode transcription factors that are known to be involved in several aspects of plant growth and development, especially in floral organ specification. To date, the comprehensive analysis of potato MADS-box gene family is still lacking after the completion of potato genome sequencing. A genome-wide characterization, classification, and expression analysis of MADS-box transcription factor gene family was performed in this study.
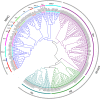
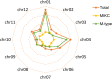
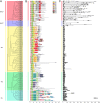
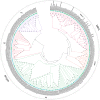
Results: A total of 153 MADS-box genes were identified and categorized into MIKC subfamily (MIKCC and MIKC*) and M-type subfamily (Mα, Mβ, and Mγ) based on their phylogenetic relationships to the Arabidopsis and rice MADS-box genes. The potato M-type subfamily had 114 members, which is almost three times of the MIKC members (39), indicating that M-type MADS-box genes have a higher duplication rate and/or a lower loss rate during potato genome evolution. Potato MADS-box genes were present on all 12 potato chromosomes with substantial clustering that mainly contributed by the M-type members. Chromosomal localization of potato MADS-box genes revealed that MADS-box genes, mostly MIKC, were located on the duplicated segments of the potato genome whereas tandem duplications mainly contributed to the M-type gene expansion. The potato MIKC subfamily could be further classified into 11 subgroups and the TT16-like, AGL17-like, and FLC-like subgroups found in Arabidopsis were absent in potato. Moreover, the expressions of potato MADS-box genes in various tissues were analyzed by using RNA-seq data and verified by quantitative real-time PCR, revealing that the MIKCC genes were mainly expressed in flower organs and several of them were highly expressed in stolon and tubers. StMADS1 and StMADS13 were up-regulated in the StSP6A-overexpression plants and down-regulated in the StSP6A-RNAi plant, and their expression in leaves and/or young tubers were associated with high level expression of StSP6A.
Conclusion: Our study identifies the family members of potato MADS-box genes and investigate the evolution history and functional divergence of MADS-box gene family. Moreover, we analyze the MIKCC expression patterns and screen for genes involved in tuberization. Finally, the StMADS1 and StMADS13 are most likely to be downstream target of StSP6A and involved in tuber development.
Keywords: MADS-box; Potato; StSP6A; Tuberigen; Tuberization.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Riechmann JL, Meyerowitz EM. MADS domain proteins in plant development. Biol Chem. 1997;378:1079–1101. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

