Efficient multi-task chemogenomics for drug specificity prediction
- PMID: 30286165
- PMCID: PMC6171913
- DOI: 10.1371/journal.pone.0204999
Efficient multi-task chemogenomics for drug specificity prediction
Abstract
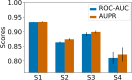
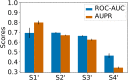
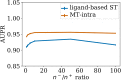
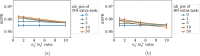
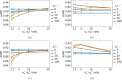
Adverse drug reactions, also called side effects, range from mild to fatal clinical events and significantly affect the quality of care. Among other causes, side effects occur when drugs bind to proteins other than their intended target. As experimentally testing drug specificity against the entire proteome is out of reach, we investigate the application of chemogenomics approaches. We formulate the study of drug specificity as a problem of predicting interactions between drugs and proteins at the proteome scale. We build several benchmark datasets, and propose NN-MT, a multi-task Support Vector Machine (SVM) algorithm that is trained on a limited number of data points, in order to solve the computational issues or proteome-wide SVM for chemogenomics. We compare NN-MT to different state-of-the-art methods, and show that its prediction performances are similar or better, at an efficient calculation cost. Compared to its competitors, the proposed method is particularly efficient to predict (protein, ligand) interactions in the difficult double-orphan case, i.e. when no interactions are previously known for the protein nor for the ligand. The NN-MT algorithm appears to be a good default method providing state-of-the-art or better performances, in a wide range of prediction scenario that are considered in the present study: proteome-wide prediction, protein family prediction, test (protein, ligand) pairs dissimilar to pairs in the train set, and orphan cases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

