LC3B is not recruited along with the autophagy elongation complex (ATG5-12/16L1) at HCV replication site and is dispensable for viral replication
- PMID: 30286180
- PMCID: PMC6171931
- DOI: 10.1371/journal.pone.0205189
LC3B is not recruited along with the autophagy elongation complex (ATG5-12/16L1) at HCV replication site and is dispensable for viral replication
Abstract
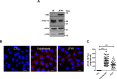
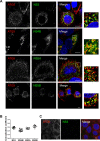
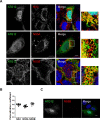
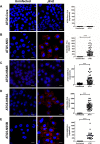
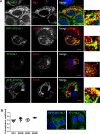
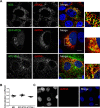
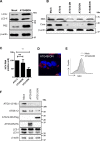
Hepatitis C virus (HCV) infection is known to induce autophagosome accumulation as observed by the typical punctate cytoplasmic distribution of LC3B-II in infected cells. Previously, we showed that viral RNA-dependent RNA polymerase (NS5B) interacts with ATG5, a major component of the autophagy elongation complex that is involved in the formation of double-membrane vesicles (DMV), and demonstrated that the autophagy elongation complex (ATG5-12/16L1) but not LC3B is required for proper membranous web formation. In this study, the colocalization and in situ interaction of all HCV replicase components with the constituent of the autophagy elongation complex and LC3B were analyzed. The results clearly show the recruitment of the elongation complex to the site of viral replication. Using in situ proximity ligation assay, we show that ATG5, but not ATG16L1, interacts with several HCV replicase components suggesting that the recruitment is directed via the ATG5-12 conjugate. Interestingly, no E3-like conjugation activity of ATG5-12/16L1 can be detected at the at HCV replication site since LC3B-II is not found along with the elongation complex at the site of viral replication. In agreement with this result, no sign of in situ interaction of LC3B with the replicase components is observed. Finally, using dominant negative forms of ATG proteins, we demonstrate that ATG5-12 conjugate, but not LC3-II formation, is critical for viral replication. Altogether, these findings suggest that although HCV needs the elongation complex for its replication, it has developed a mechanism to avoid canonical LC3-II accumulation at viral replication site.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation.J Virol. 2018 Mar 14;92(7):e01513-17. doi: 10.1128/JVI.01513-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29367244 Free PMC article.
-
The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation.Sci Rep. 2017 Jan 9;7:40351. doi: 10.1038/srep40351. Sci Rep. 2017. PMID: 28067309 Free PMC article.
-
Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection.Virology. 2010 Sep 15;405(1):1-7. doi: 10.1016/j.virol.2010.05.032. Epub 2010 Jun 26. Virology. 2010. PMID: 20580051 Free PMC article.
-
ATG5: A central autophagy regulator implicated in various human diseases.Cell Biochem Funct. 2022 Oct;40(7):650-667. doi: 10.1002/cbf.3740. Epub 2022 Sep 5. Cell Biochem Funct. 2022. PMID: 36062813 Review.
-
Impact of the autophagy machinery on hepatitis C virus infection.Viruses. 2011 Aug;3(8):1342-57. doi: 10.3390/v3081342. Epub 2011 Aug 4. Viruses. 2011. PMID: 21994783 Free PMC article. Review.
Cited by
-
Hepatitis Delta Virus Alters the Autophagy Process To Promote Its Genome Replication.J Virol. 2020 Jan 31;94(4):e01936-19. doi: 10.1128/JVI.01936-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31748400 Free PMC article.
-
Gene and protein expression of mTOR and LC3 in hepatocellular carcinoma, colorectal liver metastasis and "normal" liver tissues.PLoS One. 2020 Dec 23;15(12):e0244356. doi: 10.1371/journal.pone.0244356. eCollection 2020. PLoS One. 2020. PMID: 33362215 Free PMC article.
-
Astrovirus replication is dependent on induction of double-membrane vesicles through a PI3K-dependent, LC3-independent pathway.J Virol. 2023 Sep 28;97(9):e0102523. doi: 10.1128/jvi.01025-23. Epub 2023 Sep 5. J Virol. 2023. PMID: 37668367 Free PMC article.
-
Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation.Cell Rep. 2021 Nov 23;37(8):110049. doi: 10.1016/j.celrep.2021.110049. Epub 2021 Nov 10. Cell Rep. 2021. PMID: 34788596 Free PMC article.
References
-
- Combs C, Brunt EM, Solomon H, Bacon BR, Brantly M, Di Bisceglie AM. Rapid development of hepatic alpha1-antitrypsin globules after liver transplantation for chronic hepatitis C. Gastroenterology. 1997;112(4):1372–5. Epub 1997/04/01. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

