Lung tumor segmentation methods: Impact on the uncertainty of radiomics features for non-small cell lung cancer
- PMID: 30286184
- PMCID: PMC6171919
- DOI: 10.1371/journal.pone.0205003
Lung tumor segmentation methods: Impact on the uncertainty of radiomics features for non-small cell lung cancer
Abstract
Purpose: To evaluate the uncertainty of radiomics features from contrast-enhanced breath-hold helical CT scans of non-small cell lung cancer for both manual and semi-automatic segmentation due to intra-observer, inter-observer, and inter-software reliability.
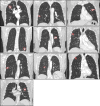
Methods: Three radiation oncologists manually delineated lung tumors twice from 10 CT scans using two software tools (3D-Slicer and MIM Maestro). Additionally, three observers without formal clinical training were instructed to use two semi-automatic segmentation tools, Lesion Sizing Toolkit (LSTK) and GrowCut, to delineate the same tumor volumes. The accuracy of the semi-automatic contours was assessed by comparison with physician manual contours using Dice similarity coefficients and Hausdorff distances. Eighty-three radiomics features were calculated for each delineated tumor contour. Informative features were identified based on their dynamic range and correlation to other features. Feature reliability was then evaluated using intra-class correlation coefficients (ICC). Feature range was used to evaluate the uncertainty of the segmentation methods.
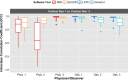
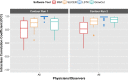
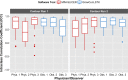
Results: From the initial set of 83 features, 40 radiomics features were found to be informative, and these 40 features were used in the subsequent analyses. For both intra-observer and inter-observer reliability, LSTK had higher reliability than GrowCut and the two manual segmentation tools. All observers achieved consistently high ICC values when using LSTK, but the ICC value varied greatly for each observer when using GrowCut and the manual segmentation tools. For inter-software reliability, features were not reproducible across the software tools for either manual or semi-automatic segmentation methods. Additionally, no feature category was found to be more reproducible than another feature category. Feature ranges of LSTK contours were smaller than those of manual contours for all features.
Conclusion: Radiomics features extracted from LSTK contours were highly reliable across and among observers. With semi-automatic segmentation tools, observers without formal clinical training were comparable to physicians in evaluating tumor segmentation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
Robust Radiomics feature quantification using semiautomatic volumetric segmentation.PLoS One. 2014 Jul 15;9(7):e102107. doi: 10.1371/journal.pone.0102107. eCollection 2014. PLoS One. 2014. PMID: 25025374 Free PMC article.
-
Assessment of liver metastases radiomic feature reproducibility with deep-learning-based semi-automatic segmentation software.Acta Radiol. 2021 Mar;62(3):291-301. doi: 10.1177/0284185120922822. Epub 2020 Jun 9. Acta Radiol. 2021. PMID: 32517533
-
The influence of image selection and segmentation on the extraction of lung cancer imaging radiomics features using 3D-Slicer software.BMC Cancer. 2025 Apr 17;25(1):728. doi: 10.1186/s12885-025-14094-z. BMC Cancer. 2025. PMID: 40247266 Free PMC article.
-
Can quantitative peritumoral CT radiomics features predict the prognosis of patients with non-small cell lung cancer? A systematic review.Eur Radiol. 2023 Mar;33(3):2105-2117. doi: 10.1007/s00330-022-09174-8. Epub 2022 Oct 29. Eur Radiol. 2023. PMID: 36307554 Free PMC article.
-
A review on multiplatform evaluations of semi-automatic open-source based image segmentation for cranio-maxillofacial surgery.Comput Methods Programs Biomed. 2019 Dec;182:105102. doi: 10.1016/j.cmpb.2019.105102. Epub 2019 Sep 30. Comput Methods Programs Biomed. 2019. PMID: 31610359 Review.
Cited by
-
Segmenting lung tumors on longitudinal imaging studies via a patient-specific adaptive convolutional neural network.Radiother Oncol. 2019 Feb;131:101-107. doi: 10.1016/j.radonc.2018.10.037. Epub 2018 Dec 31. Radiother Oncol. 2019. PMID: 30773175 Free PMC article.
-
Radiogenomics Map-Based Molecular and Imaging Phenotypical Characterization in Localised Prostate Cancer Using Pre-Biopsy Biparametric MR Imaging.Int J Mol Sci. 2024 May 15;25(10):5379. doi: 10.3390/ijms25105379. Int J Mol Sci. 2024. PMID: 38791417 Free PMC article.
-
How to read and review papers on machine learning and artificial intelligence in radiology: a survival guide to key methodological concepts.Eur Radiol. 2021 Apr;31(4):1819-1830. doi: 10.1007/s00330-020-07324-4. Epub 2020 Oct 1. Eur Radiol. 2021. PMID: 33006018 Review.
-
Performance of alternative manual and automated deep learning segmentation techniques for the prediction of benign and malignant lung nodules.J Med Imaging (Bellingham). 2023 Jul;10(4):044006. doi: 10.1117/1.JMI.10.4.044006. Epub 2023 Aug 9. J Med Imaging (Bellingham). 2023. PMID: 37564098 Free PMC article.
-
Importance of CT image normalization in radiomics analysis: prediction of 3-year recurrence-free survival in non-small cell lung cancer.Eur Radiol. 2022 Dec;32(12):8716-8725. doi: 10.1007/s00330-022-08869-2. Epub 2022 May 31. Eur Radiol. 2022. PMID: 35639142
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

