Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas
- PMID: 30286807
- PMCID: PMC6172741
- DOI: 10.1186/s40168-018-0564-7
Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas
Abstract
Background: Pediatric asthma is the most common chronic childhood disease in the USA, currently affecting ~ 7 million children. This heterogeneous syndrome is thought to encompass various disease phenotypes of clinically observable characteristics, which can be statistically identified by applying clustering approaches to patient clinical information. Extensive evidence has shown that the airway microbiome impacts both clinical heterogeneity and pathogenesis in pediatric asthma. Yet, so far, airway microbiotas have been consistently neglected in the study of asthma phenotypes. Here, we couple extensive clinical information with 16S rRNA high-throughput sequencing to characterize the microbiota of the nasal cavity in 163 children and adolescents clustered into different asthma phenotypes.
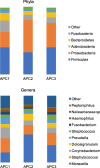
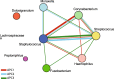
Results: Our clustering analyses identified three statistically distinct phenotypes of pediatric asthma. Four core OTUs of the pathogenic genera Moraxella, Staphylococcus, Streptococcus, and Haemophilus were present in at least 95% of the studied nasal microbiotas. Phyla (Proteobacteria, Actinobacteria, and Bacteroidetes) and genera (Moraxella, Corynebacterium, Dolosigranulum, and Prevotella) abundances, community composition, and structure varied significantly (0.05 < P ≤ 0.0001) across asthma phenotypes and one of the clinical variables (preterm birth). Similarly, microbial networks of co-occurrence of bacterial genera revealed different bacterial associations across asthma phenotypes.
Conclusions: This study shows that children and adolescents with different clinical characteristics of asthma also show different nasal bacterial profiles, which is indicative of different phenotypes of the disease. Our work also shows how clinical and microbial information could be integrated to validate and refine asthma classification systems and develop biomarkers of disease.
Keywords: 16S rRNA; Asthma; Microbiome; Nose; Phenotype.
Conflict of interest statement
Ethics approval and consent to participate
All participants in this study were part of The AsthMaP-2 Project. AsthMaP-2 and the work presented here was approved by the Children’s National Medical Center Institutional Review Board. Written consent was obtained from all independent participants or their legal guardians using the IRB-approved informed consent documents.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- National Institute of Allergy and Infectious Diseases. NIAID Strategic Plan 2013. 2013. Available from: http://www.niaid.nih.gov/about/whoweare/planningpriorities/Pages/default.... Last accessed: 6 Jan 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

