SENSITIVE TO SALT1, An Endoplasmic Reticulum-Localized Chaperone, Positively Regulates Salt Resistance
- PMID: 30287478
- PMCID: PMC6236605
- DOI: 10.1104/pp.18.00840
SENSITIVE TO SALT1, An Endoplasmic Reticulum-Localized Chaperone, Positively Regulates Salt Resistance
Abstract
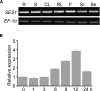
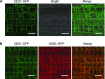
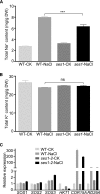
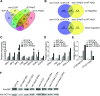
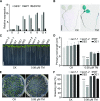
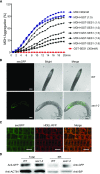
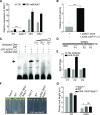
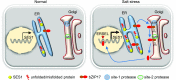
Salt stress seriously affects plant growth and development. Through genetic screening, we identified and characterized an Arabidopsis (Arabidopsis thaliana) sensitive to salt1 (ses1) mutant. SES1 was ubiquitously expressed and induced by salt treatment. The salt-sensitive phenotype of ses1 was due neither to the overaccumulation of Na+ nor to the suppression of salt tolerance-associated genes. SES1 encoded an uncharacterized endoplasmic reticulum (ER)-localized protein. Coinciding with its subcellular distribution, ses1 exhibited overactivation of unfolded protein response genes and was largely influenced by severe ER stress. Biochemical evidence revealed that SES1 functions as an important molecular chaperone to alleviate salt-induced ER stress. Furthermore, the ER stress sensor basic leucine zipper factor17 transactivated SES1 by binding directly to its promoter region. These results provide insights into salt stress responses and ER homeostasis and shed light on the mechanism by which SES1 modulates salt resistance.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


Similar articles
-
SES1 positively regulates heat stress resistance in Arabidopsis.Biochem Biophys Res Commun. 2019 Jun 4;513(3):582-588. doi: 10.1016/j.bbrc.2019.04.015. Epub 2019 Apr 10. Biochem Biophys Res Commun. 2019. PMID: 30981503
-
Endoplasmic reticulum proteins quality control and the unfolded protein response: the regulative mechanism of organisms against stress injuries.Biofactors. 2014 Nov-Dec;40(6):569-85. doi: 10.1002/biof.1194. Epub 2014 Dec 20. Biofactors. 2014. PMID: 25530003 Review.
-
Constitutive BiP protein accumulation in Arabidopsis mutants defective in a gene encoding chloroplast-resident stearoyl-acyl carrier protein desaturase.Genes Cells. 2018 Jun;23(6):456-465. doi: 10.1111/gtc.12585. Epub 2018 Apr 24. Genes Cells. 2018. PMID: 29688606
-
The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis.Plant J. 2013 Oct;76(2):274-86. doi: 10.1111/tpj.12287. Epub 2013 Aug 12. Plant J. 2013. PMID: 23869562
-
Emerging roles for diverse intramembrane proteases in plant biology.Biochim Biophys Acta. 2013 Dec;1828(12):2933-6. doi: 10.1016/j.bbamem.2013.05.013. Biochim Biophys Acta. 2013. PMID: 24099011 Review.
Cited by
-
Linear motifs regulating protein secretion, sorting and autophagy in Leishmania parasites are diverged with respect to their host equivalents.PLoS Comput Biol. 2024 Feb 16;20(2):e1011902. doi: 10.1371/journal.pcbi.1011902. eCollection 2024 Feb. PLoS Comput Biol. 2024. PMID: 38363808 Free PMC article.
-
The proteome landscape of the root cap reveals a role for the jacalin-associated lectin JAL10 in the salt-induced endoplasmic reticulum stress pathway.Plant Commun. 2023 Nov 13;4(6):100726. doi: 10.1016/j.xplc.2023.100726. Epub 2023 Oct 2. Plant Commun. 2023. PMID: 37789617 Free PMC article.
-
The Endoplasmic Reticulum Role in the Plant Response to Abiotic Stress.Front Plant Sci. 2021 Nov 18;12:755447. doi: 10.3389/fpls.2021.755447. eCollection 2021. Front Plant Sci. 2021. PMID: 34868142 Free PMC article. Review.
-
Arabidopsis Toxicos en Levadura 12 Modulates Salt Stress and ABA Responses in Arabidopsis thaliana.Int J Mol Sci. 2022 Jun 30;23(13):7290. doi: 10.3390/ijms23137290. Int J Mol Sci. 2022. PMID: 35806295 Free PMC article.
-
The thiol-disulfide exchange activity of AtPDI1 is involved in the response to abiotic stresses.BMC Plant Biol. 2021 Nov 23;21(1):557. doi: 10.1186/s12870-021-03325-7. BMC Plant Biol. 2021. PMID: 34814838 Free PMC article.
References
-
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J 49: 718–728 - PubMed
-
- Cha JY, Jung MH, Ermawati N, Su’udi M, Rho GJ, Han CD, Lee KH, Son D (2009) Functional characterization of orchardgrass endoplasmic reticulum-resident Hsp90 (DgHsp90) as a chaperone and an ATPase. Plant Physiol Biochem 47: 859–866 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

