Scrambling of natural and fluorescently tagged phosphatidylinositol by reconstituted G protein-coupled receptor and TMEM16 scramblases
- PMID: 30287690
- PMCID: PMC6254352
- DOI: 10.1074/jbc.RA118.004213
Scrambling of natural and fluorescently tagged phosphatidylinositol by reconstituted G protein-coupled receptor and TMEM16 scramblases
Abstract
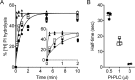
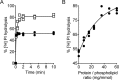
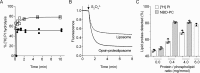
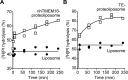
Members of the G protein-coupled receptor and TMEM16 (transmembrane protein 16) protein families are phospholipid scramblases that facilitate rapid, bidirectional movement of phospholipids across a membrane bilayer in an ATP-independent manner. On reconstitution into large unilamellar vesicles, these proteins scramble more than 10,000 lipids/protein/s as measured with co-reconstituted fluorescent nitrobenzoxadiazole (NBD)-labeled phospholipids. Although NBD-labeled phospholipids are ubiquitously used as reporters of scramblase activity, it remains unclear whether the NBD modification influences the quantitative outcomes of the scramblase assay. We now report a refined biochemical approach for measuring the activity of scramblase proteins with radiolabeled natural phosphatidylinositol ([3H]PI) and exploiting the hydrolytic activity of bacterial PI-specific phospholipase C (PI-PLC) to detect the transbilayer movement of PI. PI-PLC rapidly hydrolyzed 50% of [3H]PI in large symmetric, unilamellar liposomes, corresponding to the lipid pool in the outer leaflet. On reconstitution of a crude preparation of yeast endoplasmic reticulum scramblase, purified bovine opsin, or purified Nectria haematococca TMEM16, the extent of [3H]PI hydrolysis increased, indicating that [3H]PI from the inner leaflet had been scrambled to the outer leaflet. Using transphosphatidylation, we synthesized acyl-NBD-PI and used it to compare our PI-PLC-based assay with conventional fluorescence-based methods. Our results revealed quantitative differences between the two assays that we attribute to the specific features of the assays themselves rather than to the nature of the phospholipid. In summary, we have developed an assay that measures scrambling of a chemically unmodified phospholipid by a reconstituted scramblase.
Keywords: G protein–coupled receptor (GPCR); Phospholipase C; TMEM16; glycerophospholipid; liposome; membrane transport; phosphatidylinositol; scramblase.
© 2018 Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Structural mapping of fluorescently-tagged, functional nhTMEM16 scramblase in a lipid bilayer.J Biol Chem. 2018 Aug 3;293(31):12248-12258. doi: 10.1074/jbc.RA118.003648. Epub 2018 Jun 14. J Biol Chem. 2018. PMID: 29903908 Free PMC article.
-
Out-of-the-groove transport of lipids by TMEM16 and GPCR scramblases.Proc Natl Acad Sci U S A. 2018 Jul 24;115(30):E7033-E7042. doi: 10.1073/pnas.1806721115. Epub 2018 Jun 20. Proc Natl Acad Sci U S A. 2018. PMID: 29925604 Free PMC article.
-
Preparation of Proteoliposomes with Purified TMEM16 Protein for Accurate Measures of Lipid Scramblase Activity.Methods Mol Biol. 2019;1949:181-199. doi: 10.1007/978-1-4939-9136-5_14. Methods Mol Biol. 2019. PMID: 30790257
-
Established and emerging players in phospholipid scrambling: A structural perspective.Biochimie. 2024 Dec;227(Pt B):111-122. doi: 10.1016/j.biochi.2024.09.008. Epub 2024 Sep 18. Biochimie. 2024. PMID: 39304020 Review.
-
Wide-ranging cellular functions of ion channels and lipid scramblases in the structurally related TMC, TMEM16 and TMEM63 families.Nat Struct Mol Biol. 2025 Feb;32(2):222-236. doi: 10.1038/s41594-024-01444-x. Epub 2024 Dec 23. Nat Struct Mol Biol. 2025. PMID: 39715905 Review.
Cited by
-
Genome-wide CRISPR screen reveals CLPTM1L as a lipid scramblase required for efficient glycosylphosphatidylinositol biosynthesis.Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2115083119. doi: 10.1073/pnas.2115083119. Epub 2022 Mar 28. Proc Natl Acad Sci U S A. 2022. PMID: 35344438 Free PMC article.
-
Lipid Dyshomeostasis and Inherited Cerebellar Ataxia.Mol Neurobiol. 2022 Jun;59(6):3800-3828. doi: 10.1007/s12035-022-02826-2. Epub 2022 Apr 14. Mol Neurobiol. 2022. PMID: 35420383 Free PMC article. Review.
-
Phospholipids are imported into mitochondria by VDAC, a dimeric beta barrel scramblase.Nat Commun. 2023 Dec 8;14(1):8115. doi: 10.1038/s41467-023-43570-y. Nat Commun. 2023. PMID: 38065946 Free PMC article.
-
TMEM16 scramblases thin the membrane to enable lipid scrambling.Nat Commun. 2022 May 11;13(1):2604. doi: 10.1038/s41467-022-30300-z. Nat Commun. 2022. PMID: 35562175 Free PMC article.
-
Lactose Permease Scrambles Phospholipids.Biology (Basel). 2023 Oct 25;12(11):1367. doi: 10.3390/biology12111367. Biology (Basel). 2023. PMID: 37997967 Free PMC article.
References
-
- Kennedy E. P., and Weiss S. B. (1956) The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222, 193–214 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

