Discovery of an MLLT1/3 YEATS Domain Chemical Probe
- PMID: 30288907
- PMCID: PMC6348381
- DOI: 10.1002/anie.201810617
Discovery of an MLLT1/3 YEATS Domain Chemical Probe
Abstract
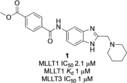
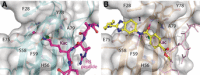
YEATS domain (YD) containing proteins are an emerging class of epigenetic targets in drug discovery. Dysregulation of these modified lysine-binding proteins has been linked to the onset and progression of cancers. We herein report the discovery and characterisation of the first small-molecule chemical probe, SGC-iMLLT, for the YD of MLLT1 (ENL/YEATS1) and MLLT3 (AF9/YEATS3). SGC-iMLLT is a potent and selective inhibitor of MLLT1/3-histone interactions. Excellent selectivity over other human YD proteins (YEATS2/4) and bromodomains was observed. Furthermore, our probe displays cellular target engagement of MLLT1 and MLLT3. The first small-molecule X-ray co-crystal structures with the MLLT1 YD are also reported. This first-in-class probe molecule can be used to understand MLLT1/3-associated biology and the therapeutic potential of small-molecule YD inhibitors.
Keywords: MLLT1; MLLT3; YEATS; chemical probes; epigenetics.
© 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
N. Manevski and J. Heer are employees and hold shares in UCB Pharma Ltd.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

