Deep learning and virtual drug screening
- PMID: 30288997
- PMCID: PMC6563286
- DOI: 10.4155/fmc-2018-0314
Deep learning and virtual drug screening
Abstract
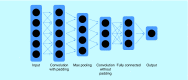
Current drug development is still costly and slow given tremendous technological advancements in drug discovery and medicinal chemistry. Using machine learning (ML) to virtually screen compound libraries promises to fix this for generating drug leads more efficiently and accurately. Herein, we explain the broad basics and integration of both virtual screening (VS) and ML. We then discuss artificial neural networks (ANNs) and their usage for VS. The ANN is emerging as the dominant classifier for ML in general, and has proven its utility for both structure-based and ligand-based VS. Techniques such as dropout, multitask learning and convolution improve the performance of ANNs and enable them to take on chemical meaning when learning about the drug-target-binding activity of compounds.
Keywords: artificial intelligence; artificial neural networks; convolutional neural networks; deep learning; drug discovery; machine learning; multitask learning; virtual screening.
Conflict of interest statement
This work was partially supported by a NIH grant R01AG056614 (to X Huang). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures

References
-
- Dimasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016;47:20–33. - PubMed
-
- Pitt WR, Calmiano MD, Kroeplien B, Taylor RD, Turner JP, King MA. Structure-based virtual screening for novel ligands. In: Williams MA, Daviter T, editors. Protein-Ligand Interactions: Methods and Applications. Humana Press; Totowa, NJ, USA: 2013. pp. 501–519. - PubMed
-
- Cereto-Massague A, Ojeda MJ, Valls C, Mulero M, Garcia-Vallve S, Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. - PubMed
-
- Molecular Operating Environment (MOE) 2018. www.chemcomp.com/MOE-Molecular_Operating_Environment.htm
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
