CellMarker: a manually curated resource of cell markers in human and mouse
- PMID: 30289549
- PMCID: PMC6323899
- DOI: 10.1093/nar/gky900
CellMarker: a manually curated resource of cell markers in human and mouse
Abstract
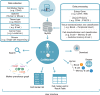
One of the most fundamental questions in biology is what types of cells form different tissues and organs in a functionally coordinated fashion. Larger-scale single-cell sequencing and biology experiment studies are now rapidly opening up new ways to track this question by revealing substantial cell markers for distinguishing different cell types in tissues. Here, we developed the CellMarker database (http://biocc.hrbmu.edu.cn/CellMarker/ or http://bio-bigdata.hrbmu.edu.cn/CellMarker/), aiming to provide a comprehensive and accurate resource of cell markers for various cell types in tissues of human and mouse. By manually curating over 100 000 published papers, 4124 entries including the cell marker information, tissue type, cell type, cancer information and source, were recorded. At last, 13 605 cell markers of 467 cell types in 158 human tissues/sub-tissues and 9148 cell makers of 389 cell types in 81 mouse tissues/sub-tissues were collected and deposited in CellMarker. CellMarker provides a user-friendly interface for browsing, searching and downloading markers of diverse cell types of different tissues. Furthermore, a summarized marker prevalence in each cell type is graphically and intuitively presented through a vivid statistical graph. We believe that CellMarker is a comprehensive and valuable resource for cell researches in precisely identifying and characterizing cells, especially at the single-cell level.
Figures



References
-
- Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K., Wolf D., Saliba A.E., Zernecke A.. Single-Cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 2018; 122:1661–1674. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

