Myeloid TBK1 Signaling Contributes to the Immune Response to Influenza
- PMID: 30290124
- PMCID: PMC6397979
- DOI: 10.1165/rcmb.2018-0122OC
Myeloid TBK1 Signaling Contributes to the Immune Response to Influenza
Abstract
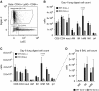
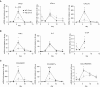
Macrophages provide key elements of the host response to influenza A virus (IAV) infection, including expression of type I IFN and inflammatory cytokines and chemokines. TBK1 (TNF receptor-associated factor family member-associated NF-κB activator-binding kinase 1) contributes to IFN expression and antiviral responses in some cell types, but its role in the innate response to IAV in vivo is unknown. We hypothesized that macrophage TBK1 contributes to both IFN and non-IFN components of host defense and IAV pathology. We generated myeloid-conditional TBK1 knockout mice and assessed the in vitro and in vivo consequences of IAV infection. Myeloid-specific loss of TBK1 in vivo resulted in less severe host response to IAV, as assessed by decreased mortality, weight loss, and hypoxia and less inflammatory changes in BAL fluid relative to wild-type mice despite no differences in viral load. Mice lacking myeloid TBK1 showed less recruitment of CD64+SiglecF-Ly6Chi inflammatory macrophages, less expression of inflammatory cytokines in the BAL fluid, and less expression of both IFN regulatory factor and NF-κB target genes in the lung. Analysis of sorted alveolar macrophages, inflammatory macrophages, and lung interstitial macrophages revealed that each subpopulation requires TBK1 for distinct components of the response to IAV infection. Our findings define roles for myeloid TBK1 in IAV-induced lung inflammation apart from IFN type I expression and point to myeloid TBK1 as a central and cell type-specific regulator of virus-induced lung damage.
Keywords: IFN; TBK1; cytokine; influenza; macrophage.
Figures





Comment in
-
TANKing Influenza A Virus in the Lung.Am J Respir Cell Mol Biol. 2019 Mar;60(3):255-256. doi: 10.1165/rcmb.2018-0337ED. Am J Respir Cell Mol Biol. 2019. PMID: 30365353 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

