From APOBEC to ZAP: Diverse mechanisms used by cellular restriction factors to inhibit virus infections
- PMID: 30290238
- PMCID: PMC6334645
- DOI: 10.1016/j.bbamcr.2018.09.012
From APOBEC to ZAP: Diverse mechanisms used by cellular restriction factors to inhibit virus infections
Abstract
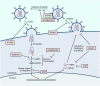
Antiviral restriction factors are cellular proteins that inhibit the entry, replication, or spread of viruses. These proteins are critical components of the innate immune system and function to limit the severity and host range of virus infections. Here we review the current knowledge on the mechanisms of action of several restriction factors that affect multiple viruses at distinct stages of their life cycles. For example, APOBEC3G deaminates cytosines to hypermutate reverse transcribed viral DNA; IFITM3 alters membranes to inhibit virus membrane fusion; MXA/B oligomerize on viral protein complexes to inhibit virus replication; SAMHD1 decreases dNTP intracellular concentrations to prevent reverse transcription of retrovirus genomes; tetherin prevents release of budding virions from cells; Viperin catalyzes formation of a nucleoside analogue that inhibits viral RNA polymerases; and ZAP binds virus RNAs to target them for degradation. We also discuss countermeasures employed by specific viruses against these restriction factors, and mention secondary functions of several of these factors in modulating immune responses. These important examples highlight the diverse strategies cells have evolved to combat virus infections.
Keywords: Interferon stimulated gene; Restriction factor; Virus infection.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures

References
-
- Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., Dabelic R., Manicassamy B., Aitchison J.D., Aderem A., Elliott R.M., Garcia-Sastre A., Racaniello V., Snijder E.J., Yokoyama W.M., Diamond M.S., Virgin H.W., Rice C.M. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. - PMC - PubMed
-
- Daugherty M.D., Malik H.S. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46(46):677–700. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

