A rapid rate of sex-chromosome turnover and non-random transitions in true frogs
- PMID: 30291233
- PMCID: PMC6173717
- DOI: 10.1038/s41467-018-06517-2
A rapid rate of sex-chromosome turnover and non-random transitions in true frogs
Abstract
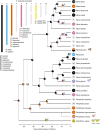
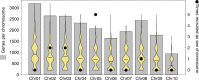
The canonical model of sex-chromosome evolution predicts that, as recombination is suppressed along sex chromosomes, gametologs will progressively differentiate, eventually becoming heteromorphic. However, there are numerous examples of homomorphic sex chromosomes across the tree of life. This homomorphy has been suggested to result from frequent sex-chromosome turnovers, yet we know little about which forces drive them. Here, we describe an extremely fast rate of turnover among 28 species of Ranidae. Transitions are not random, but converge on several chromosomes, potentially due to genes they harbour. Transitions also preserve the ancestral pattern of male heterogamety, in line with the 'hot-potato' model of sex-chromosome transitions, suggesting a key role for mutation-load accumulation in non-recombining genomic regions. The importance of mutation-load selection in frogs might result from the extreme heterochiasmy they exhibit, making frog sex chromosomes differentiate immediately from emergence and across their entire length.
Conflict of interest statement
The authors declare no competing interests.
Figures