Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery
- PMID: 30291234
- PMCID: PMC6173783
- DOI: 10.1038/s41467-018-06411-x
Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery
Abstract
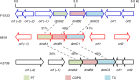
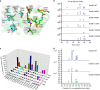
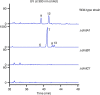
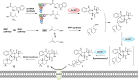
Cyclodipeptide synthases (CDPSs) can catalyze the formation of two successive peptide bonds by hijacking aminoacyl-tRNAs from the ribosomal machinery resulting in diketopiperazines (DKPs). Here, three CDPS-containing loci (dmt1-3) are discovered by genome mining and comparative genome analysis of Streptomyces strains. Among them, CDPS DmtB1, encoded by the gene of dmt1 locus, can synthesize cyclo(L-Trp-L-Xaa) (with Xaa being Val, Pro, Leu, Ile, or Ala). Systematic mutagenesis experiments demonstrate the importance of the residues constituting substrate-binding pocket P1 for the incorporation of the second aa-tRNA in DmtB1. Characterization of dmt1-3 unravels that CDPS-dependent machinery is involved in CDPS-synthesized DKP formation followed by tailoring steps of prenylation and cyclization to afford terpenylated DKP compounds drimentines. A phytoene-synthase-like family prenyltransferase (DmtC1) and a membrane terpene cyclase (DmtA1) are required for drimentines biosynthesis. These results set the foundation for further increasing the natural diversity of complex DKP derivatives.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Deciphering a Cyclodipeptide Synthase Pathway Encoding Prenylated Indole Alkaloids in Streptomyces leeuwenhoekii.Appl Environ Microbiol. 2021 May 11;87(11):e02525-20. doi: 10.1128/AEM.02525-20. Print 2021 May 11. Appl Environ Microbiol. 2021. PMID: 33741615 Free PMC article.
-
The expanding spectrum of diketopiperazine natural product biosynthetic pathways containing cyclodipeptide synthases.Org Biomol Chem. 2019 Feb 27;17(9):2305-2314. doi: 10.1039/c8ob03063d. Org Biomol Chem. 2019. PMID: 30688950 Review.
-
Expanding tryptophan-containing cyclodipeptide synthase spectrum by identification of nine members from Streptomyces strains.Appl Microbiol Biotechnol. 2018 May;102(10):4435-4444. doi: 10.1007/s00253-018-8908-6. Epub 2018 Mar 24. Appl Microbiol Biotechnol. 2018. PMID: 29574613
-
Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes.Nat Chem Biol. 2009 Jun;5(6):414-20. doi: 10.1038/nchembio.175. Nat Chem Biol. 2009. PMID: 19430487
-
The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways.Nat Prod Rep. 2012 Sep;29(9):961-79. doi: 10.1039/c2np20010d. Epub 2012 Jul 3. Nat Prod Rep. 2012. PMID: 22751625 Review.
Cited by
-
Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.J Am Chem Soc. 2020 Oct 14;142(41):17413-17424. doi: 10.1021/jacs.0c06312. Epub 2020 Sep 30. J Am Chem Soc. 2020. PMID: 32786740 Free PMC article.
-
Expanding the Structural Diversity of Drimentines by Exploring the Promiscuity of Two N-methyltransferases.iScience. 2020 Jul 24;23(7):101323. doi: 10.1016/j.isci.2020.101323. Epub 2020 Jun 28. iScience. 2020. PMID: 32659721 Free PMC article.
-
Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source.Molecules. 2020 May 29;25(11):2546. doi: 10.3390/molecules25112546. Molecules. 2020. PMID: 32486092 Free PMC article.
-
Research Progress of the Biosynthesis of Natural Bio-Antibacterial Agent Pulcherriminic Acid in Bacillus.Molecules. 2020 Nov 28;25(23):5611. doi: 10.3390/molecules25235611. Molecules. 2020. PMID: 33260656 Free PMC article. Review.
-
Terpene synthases in disguise: enzymology, structure, and opportunities of non-canonical terpene synthases.Nat Prod Rep. 2020 Mar 25;37(3):425-463. doi: 10.1039/c9np00051h. Nat Prod Rep. 2020. PMID: 31650156 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

