Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88
- PMID: 30291253
- PMCID: PMC6173721
- DOI: 10.1038/s41467-018-06658-4
Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88
Erratum in
-
Author Correction: Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88.Nat Commun. 2019 Jan 28;10(1):526. doi: 10.1038/s41467-019-08456-y. Nat Commun. 2019. PMID: 30692545 Free PMC article.
Abstract
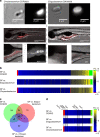
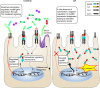
Host pathways mediating changes in immune states elicited by intestinal microbial colonization are incompletely characterized. Here we describe alterations of the host immune state induced by colonization of germ-free zebrafish larvae with an intestinal microbial community or single bacterial species. We show that microbiota-induced changes in intestinal leukocyte subsets and whole-body host gene expression are dependent on the innate immune adaptor gene myd88. Similar patterns of gene expression are elicited by colonization with conventional microbiome, as well as mono-colonization with two different zebrafish commensal bacterial strains. By studying loss-of-function myd88 mutants, we find that colonization suppresses Myd88 at the mRNA level. Tlr2 is essential for microbiota-induced effects on myd88 transcription and intestinal immune cell composition.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

