Structural basis for σ1 receptor ligand recognition
- PMID: 30291362
- PMCID: PMC6261271
- DOI: 10.1038/s41594-018-0137-2
Structural basis for σ1 receptor ligand recognition
Abstract
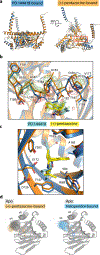
The σ1 receptor is a poorly understood membrane protein expressed throughout the human body. Ligands targeting the σ1 receptor are in clinical trials for treatment of Alzheimer's disease, ischemic stroke, and neuropathic pain. However, relatively little is known regarding the σ1 receptor's molecular function. Here, we present crystal structures of human σ1 receptor bound to the antagonists haloperidol and NE-100, and the agonist (+)-pentazocine, at crystallographic resolutions of 3.1 Å, 2.9 Å, and 3.1 Å, respectively. These structures reveal a unique binding pose for the agonist. The structures and accompanying molecular dynamics (MD) simulations identify agonist-induced structural rearrangements in the receptor. Additionally, we show that ligand binding to σ1 is a multistep process that is rate limited by receptor conformational change. We used MD simulations to reconstruct a ligand binding pathway involving two major conformational changes. These data provide a framework for understanding the molecular basis for σ1 agonism.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures


References
-
- Martin WR, Eades CG, Thompson JA, Huppler RE & Gilbert PE The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther 197, 517–532 (1976). - PubMed
-
- Su TP Evidence for sigma opioid receptor: binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J Pharmacol Exp Ther 223, 284–290 (1982). - PubMed
-
- Hellewell SB & Bowen WD A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res 527, 244–253 (1990). - PubMed
Methods-only References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

