Mycobacteriophages
- PMID: 30291704
- PMCID: PMC6282025
- DOI: 10.1128/microbiolspec.GPP3-0026-2018
Mycobacteriophages
Abstract
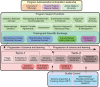
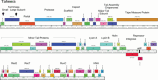
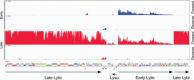
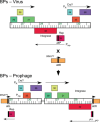
Mycobacteriophages are viruses that infect mycobacterial hosts. A large number of mycobacteriophages have been isolated and genomically characterized, providing insights into viral diversity and evolution, as well as fueling development of tools for mycobacterial genetics. Mycobacteriophages have intimate relationships with their hosts and provide insights into the genetics and physiology of the mycobacteria and tools for potential clinical applications such as drug development, diagnosis, vaccines, and potentially therapy.
Figures




References
-
- Grange JM. 1975. Proceedings: bacteriophage typing of strains of Mycobacterium tuberculosis isolated in south-east England. J Med Microbiol 8:ix. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

