A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity
- PMID: 30291794
- PMCID: PMC6246790
- DOI: 10.1002/anie.201806901
A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity
Abstract
Extracellular vesicles (EVs) actively participate in intercellular communication and pathological processes. Studying the molecular signatures of EVs is key to reveal their biological functions and clinical values, which, however, is greatly hindered by their sub-100 nm dimensions, the low quantities of biomolecules each EV carries, and the large population heterogeneity. Now, single-EV flow cytometry analysis is introduced to realize single EV counting and phenotyping in a conventional flow cytometer for the first time, enabled by target-initiated engineering (TIE) of DNA nanostructures on each EV. By illuminating multiple markers on single EVs, statistically significant differences are revealed among the molecular signatures of EVs originating from several breast cancer cell lines, and the cancer cell-derived EVs among the heterogeneous EV populations are successfully recognized. Thus, our approach holds great potential for various biological and biomedical applications.
Keywords: engineering; flow cytometry analysis; heterogeneity; molecular signatures; single extracellular vesicle analysis.
© 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


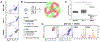
References
-
- Tkach M, Thery C, Cell 2016, 164, 1226–1232. - PubMed
-
- Jiang Y, Shi M, Liu Y, Wan S, Cui C, Zhang L, Tan W, Angew. Chem. Int. Ed. 2017, 56, 11916–11920. - PMC - PubMed
- Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ, Methods Mol. Biol. 2015, 1295, 179–209. - PubMed
- Pocsfalvi G, Stanly C, Fiume I, Vekey K, Chromatogr JA 2016, 1439, 26–41. - PubMed
- Zhao Z, Yang Y, Zeng Y, He M, Lab Chip 2016, 16, 489–496. - PMC - PubMed
- Rider MA, Hurwitz SN, Meckes DG, Sci. Rep. 2016, 6, 23978. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

