Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP
- PMID: 30293838
- PMCID: PMC6219920
- DOI: 10.1016/j.devcel.2018.09.003
Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP
Abstract
Centromeric chromatin defines the site of kinetochore formation and ensures faithful chromosome segregation. Centromeric identity is epigenetically specified by the incorporation of CENP-A nucleosomes. DNA replication presents a challenge for inheritance of centromeric identity because nucleosomes are removed to allow for replication fork progression. Despite this challenge, CENP-A nucleosomes are stably retained through S phase. We used BioID to identify proteins transiently associated with CENP-A during DNA replication. We found that during S phase, HJURP transiently associates with centromeres and binds to pre-existing CENP-A, suggesting a distinct role for HJURP in CENP-A retention. We demonstrate that HJURP is required for centromeric nucleosome inheritance during S phase. HJURP co-purifies with the MCM2-7 helicase complex and, together with the MCM2 subunit, binds CENP-A simultaneously. Therefore, pre-existing CENP-A nucleosomes require an S phase function of the HJURP chaperone and interaction with MCM2 to ensure faithful inheritance of centromere identity through DNA replication.
Keywords: DNA replication; centromere; chromatin; chromosome; epigenetics; helicase; mitosis; nucleosome.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures
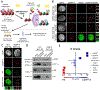



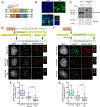
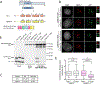

Comment in
-
Chromosomes: Keeping Centromeric Chromatin Tidy through S Phase.Curr Biol. 2019 Jan 7;29(1):R35-R37. doi: 10.1016/j.cub.2018.11.044. Curr Biol. 2019. PMID: 30620916
Similar articles
-
Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP.Cell. 2009 May 1;137(3):472-84. doi: 10.1016/j.cell.2009.02.039. Cell. 2009. PMID: 19410544 Free PMC article.
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
-
Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition.EMBO J. 2013 Jul 31;32(15):2113-24. doi: 10.1038/emboj.2013.142. Epub 2013 Jun 14. EMBO J. 2013. PMID: 23771058 Free PMC article.
-
HJURP interaction with the condensin II complex during G1 promotes CENP-A deposition.Mol Biol Cell. 2017 Jan 1;28(1):54-64. doi: 10.1091/mbc.E15-12-0843. Epub 2016 Nov 2. Mol Biol Cell. 2017. PMID: 27807043 Free PMC article.
-
Orchestrating the Specific Assembly of Centromeric Nucleosomes.Prog Mol Subcell Biol. 2017;56:165-192. doi: 10.1007/978-3-319-58592-5_7. Prog Mol Subcell Biol. 2017. PMID: 28840237 Free PMC article. Review.
Cited by
-
The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle.Open Biol. 2020 Jun;10(6):200051. doi: 10.1098/rsob.200051. Epub 2020 Jun 10. Open Biol. 2020. PMID: 32516549 Free PMC article. Review.
-
DNAJC9 prevents CENP-A mislocalization and chromosomal instability by maintaining the fidelity of histone supply chains.EMBO J. 2024 Jun;43(11):2166-2197. doi: 10.1038/s44318-024-00093-6. Epub 2024 Apr 10. EMBO J. 2024. PMID: 38600242 Free PMC article.
-
Solid tumours hijack the histone variant network.Nat Rev Cancer. 2021 Apr;21(4):257-275. doi: 10.1038/s41568-020-00330-0. Epub 2021 Feb 10. Nat Rev Cancer. 2021. PMID: 33568791 Free PMC article. Review.
-
Unveiling the mysteries of extrachromosomal circular DNA: from generation to clinical relevance in human cancers and health.Mol Cancer. 2024 Dec 20;23(1):276. doi: 10.1186/s12943-024-02187-5. Mol Cancer. 2024. PMID: 39707444 Free PMC article. Review.
-
CENP-A Regulation and Cancer.Front Cell Dev Biol. 2022 Jun 2;10:907120. doi: 10.3389/fcell.2022.907120. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35721491 Free PMC article. Review.
References
-
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, and Groth A (2014). Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16, 281–293. - PMC - PubMed
-
- Alabert C, and Groth A (2012). Chromatin replication and epigenome maintenance. Nature reviews Molecular cell biology 13, 153–167. - PubMed
-
- Annunziato AT, Schindler RK, Thomas CA Jr., and Seale RL (1981). Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. The Journal of biological chemistry 256, 11880–11886. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

