Occurrence of Highly Conjugative IncX3 Epidemic Plasmid Carrying bla NDM in Enterobacteriaceae Isolates in Geographically Widespread Areas
- PMID: 30294321
- PMCID: PMC6158458
- DOI: 10.3389/fmicb.2018.02272
Occurrence of Highly Conjugative IncX3 Epidemic Plasmid Carrying bla NDM in Enterobacteriaceae Isolates in Geographically Widespread Areas
Abstract
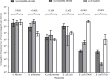
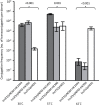
The emergence of New Delhi metallo-β-lactamase (NDM) in common enterobacterial species is a major concern for healthcare. Early reports have revealed that the spread of NDM involved diverse and heterogeneous plasmids. Recently, the involvement of a rare, IncX3 subtype plasmid has been increasingly recognized. Here, we studied the prevalence of IncX plasmid subtypes in 198 carbapenem-resistant Enterobacteriaceae, originating from a territory-wide active surveillance in Hong Kong in 2016. The complete sequences and biological features of the bla NDM-carrying plasmids were investigated. A total of 62 NDM-type, 21 OXA-48 type, 14 IMP-type, 8 KPC-type, 4 IMI-type producers, and 89 non-carbapenemase-producers were tested for presence of IncX subtypes. IncX3 (n = 60) was the most common subtype, followed by IncX4 (n = 6) and IncX1 (n = 2). The prevalence of IncX3 subtype in isolates producing NDM, other carbapenemase types and non-carbapenemase producers were 75.8, 21.3, and 3.4%, respectively (P < 0.001). An IncX3 plasmid (size ∼50 kb) was confirmed to carry bla NDM in 47 isolates of different enterobacterial species. Thirteen IncX3 plasmids originating from six healthcare regions in Hong Kong were completely sequenced. The results showed that the IncX3 plasmids carrying bla NDM share a high degree of sequence identity with a previously reported plasmid, pNDM-HN380 (GenBank accession JX104760), over the backbone and genetic load regions. A blast search further revealed the occurrence of identical or nearly identical IncX3 plasmids carrying bla NDM in other part of China, Korea, Myanmar, India, Oman, Kuwait, Italy, and Canada. Two IncX3 carrying bla NDM were investigated further. Conjugation experiments demonstrated that the IncX3 plasmids could be efficiently transferred to multiple enterobacterial species at frequencies that are comparable or higher than the epidemic IncFII plasmid carrying bla CTX-M (pHK01). In addition, efficient transfer of the NDM plasmids occurred over a range of temperatures. In conclusion, this study demonstrated the important role played by IncX3 in the dissemination of NDM and the occurrence of pNDM-HN380-like plasmids in geographically widespread areas. The high mobility of IncX3 plasmid across different enterobacterial species highlights the ability of this plasmid replicon to be an important vehicle in worldwide dissemination of NDM.
Keywords: Enterobacteriaceae; antimicrobial resistance epidemiology; carbapenems; molecular epidemiology; resistance plasmid.
Figures

References
-
- Chen J. H., Ho P. L., Kwan G. S., She K. K., Siu G. K., Cheng V. C., et al. (2013). Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J. Clin. Microbiol. 51 1733–1739. 10.1128/JCM.03259-12 - DOI - PMC - PubMed
-
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S28. Wayne, PA: CLSI.
LinkOut - more resources
Full Text Sources
Research Materials

