Deconstructing deployment of the innate immune lymphocyte army for barrier homeostasis and protection
- PMID: 30294966
- PMCID: PMC6446816
- DOI: 10.1111/imr.12709
Deconstructing deployment of the innate immune lymphocyte army for barrier homeostasis and protection
Abstract
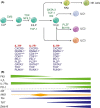
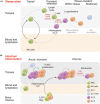
The study of the immune system has shifted from a purely dichotomous separation between the innate and adaptive arms to one that is now highly complex and reshaping our ideas of how steady-state health is assured. It is now clear that immune cells do not neatly fit into these two streams and immune homeostasis depends on continual dialogue between multiple lineages of the innate (including dendritic cells, innate lymphoid cells, and unconventional lymphocytes) and adaptive (T and B lymphocytes) arms together with a finely tuned synergy between the host and microbes which is essential to ensure immune homeostasis. Innate lymphoid cells are critical players in this new landscape. Here, we discuss recent studies that have elucidated in detail the development of ILCs from their earliest progenitors and examine factors that influence their identification and ability to drive immune homeostasis and long-term immune protection.
Keywords: homeostasis; innate immunity; innate lymphoid cells; mucosal immunity; protection.
© 2018 The Authors. Immunological Reviews Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Innate Lymphoid Cells: Regulators of Gut Barrier Function and Immune Homeostasis.J Immunol Res. 2019 Dec 20;2019:2525984. doi: 10.1155/2019/2525984. eCollection 2019. J Immunol Res. 2019. PMID: 31930146 Free PMC article. Review.
-
Control of pathogens and microbiota by innate lymphoid cells.Microbes Infect. 2018 Jun-Jul;20(6):317-322. doi: 10.1016/j.micinf.2018.05.003. Epub 2018 May 28. Microbes Infect. 2018. PMID: 29852240 Review.
-
Shared Transcriptional Control of Innate Lymphoid Cell and Dendritic Cell Development.Annu Rev Cell Dev Biol. 2019 Oct 6;35:381-406. doi: 10.1146/annurev-cellbio-100818-125403. Epub 2019 Jul 5. Annu Rev Cell Dev Biol. 2019. PMID: 31283378 Free PMC article. Review.
-
Complementary diversification of dendritic cells and innate lymphoid cells.Curr Opin Immunol. 2014 Aug;29:69-78. doi: 10.1016/j.coi.2014.04.006. Epub 2014 May 27. Curr Opin Immunol. 2014. PMID: 24874447 Free PMC article. Review.
-
The interaction of intestinal microbiota and innate lymphoid cells in health and disease throughout life.Immunology. 2020 Jan;159(1):39-51. doi: 10.1111/imm.13138. Epub 2019 Nov 27. Immunology. 2020. PMID: 31777064 Free PMC article. Review.
Cited by
-
Innate Lymphoid Cells in Colorectal Cancers: A Double-Edged Sword.Front Immunol. 2020 Jan 15;10:3080. doi: 10.3389/fimmu.2019.03080. eCollection 2019. Front Immunol. 2020. PMID: 32010138 Free PMC article. Review.
-
Immune Checkpoints and Innate Lymphoid Cells-New Avenues for Cancer Immunotherapy.Cancers (Basel). 2021 Nov 27;13(23):5967. doi: 10.3390/cancers13235967. Cancers (Basel). 2021. PMID: 34885076 Free PMC article. Review.
-
Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives.Int J Mol Sci. 2021 Aug 22;22(16):9044. doi: 10.3390/ijms22169044. Int J Mol Sci. 2021. PMID: 34445750 Free PMC article. Review.
-
Type 1 Innate Lymphoid Cells Are Proinflammatory Effector Cells in Ischemia-Reperfusion Injury of Steatotic Livers.Front Immunol. 2022 Jun 27;13:899525. doi: 10.3389/fimmu.2022.899525. eCollection 2022. Front Immunol. 2022. PMID: 35833123 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

