Identification of new high affinity targets for Roquin based on structural conservation
- PMID: 30295819
- PMCID: PMC6294493
- DOI: 10.1093/nar/gky908
Identification of new high affinity targets for Roquin based on structural conservation
Abstract
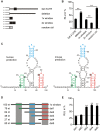
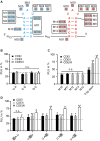
Post-transcriptional gene regulation controls the amount of protein produced from a specific mRNA by altering both its decay and translation rates. Such regulation is primarily achieved by the interaction of trans-acting factors with cis-regulatory elements in the untranslated regions (UTRs) of mRNAs. These interactions are guided either by sequence- or structure-based recognition. Similar to sequence conservation, the evolutionary conservation of a UTR's structure thus reflects its functional importance. We used such structural conservation to identify previously unknown cis-regulatory elements. Using the RNA folding program Dynalign, we scanned all UTRs of humans and mice for conserved structures. Characterizing a subset of putative conserved structures revealed a binding site of the RNA-binding protein Roquin. Detailed functional characterization in vivo enabled us to redefine the binding preferences of Roquin and identify new target genes. Many of these new targets are unrelated to the established role of Roquin in inflammation and immune responses and thus highlight additional, unstudied cellular functions of this important repressor. Moreover, the expression of several Roquin targets is highly cell-type-specific. In consequence, these targets are difficult to detect using methods dependent on mRNA abundance, yet easily detectable with our unbiased strategy.
Figures



References
-
- Lee F.C.Y., Ule J.. Advances in CLIP technologies for studies of protein-RNA interactions. Mol. Cell. 2018; 69:354–369. - PubMed
-
- Ding Y., Kwok C.K., Tang Y., Bevilacqua P.C., Assmann S.M.. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat. Protoc. 2015; 10:1050–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

