Effect of an Intrinsically Disordered Plant Stress Protein on the Properties of Water
- PMID: 30297135
- PMCID: PMC6224673
- DOI: 10.1016/j.bpj.2018.09.014
Effect of an Intrinsically Disordered Plant Stress Protein on the Properties of Water
Abstract
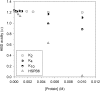
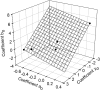
Dehydrins are plant proteins that are able to protect plants from various forms of dehydrative stress such as drought, cold, and high salinity. Dehydrins can prevent enzymes from losing activity after freeze/thaw treatments. Previous studies had suggested that the dehydrins function by a molecular shield effect, essentially preventing a denatured enzyme from aggregating with another enzyme. Therefore, the larger the dehydrin, the larger the shield and theoretically the more effective the protection. Although this relationship holds for smaller dehydrins, it fails to explain why larger dehydrins are less efficient than would be predicted from their size. Using solvatochromic dyes to probe the solvent features of water, we first confirm that the dehydrins do not bind the dyes, which would interfere with interpretation of the data. We then show that the dehydrins have an effect on three solvent properties of water (dipolarity/polarizability, hydrogen-bond donor acidity and hydrogen-bond acceptor basicity), which can contribute to the protective mechanism of these proteins. Interpretation of these data suggests that although polyethylene glycol and dehydrins have similar protective effects, dehydrins may more efficiently modify the hydrogen-bonding ability of bulk water to prevent enzyme denaturation. This possibly explains why dehydrins recover slightly more enzyme activity than polyethylene glycol.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. - PubMed
-
- Uversky V.N., Gillespie J.R., Fink A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. - PubMed
-
- Dunker A.K., Lawson J.D., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. - PubMed
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

