Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer
- PMID: 30297358
- PMCID: PMC6328329
- DOI: 10.1158/2159-8290.CD-18-0689
Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer
Abstract
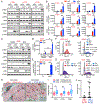
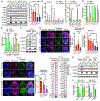
KRAS-driven lung cancers frequently inactivate TP53 and/or STK11/LKB1, defining tumor subclasses with emerging clinical relevance. Specifically, KRAS-LKB1 (KL)-mutant lung cancers are particularly aggressive, lack PD-L1, and respond poorly to immune checkpoint blockade (ICB). The mechanistic basis for this impaired immunogenicity, despite the overall high mutational load of KRAS-mutant lung cancers, remains obscure. Here, we report that LKB1 loss results in marked silencing of stimulator of interferon genes (STING) expression and insensitivity to cytoplasmic double-strand DNA (dsDNA) sensing. This effect is mediated at least in part by hyperactivation of DNMT1 and EZH2 activity related to elevated S-adenylmethionine levels and reinforced by DNMT1 upregulation. Ectopic expression of STING in KL cells engages IRF3 and STAT1 signaling downstream of TBK1 and impairs cellular fitness, due to the pathologic accumulation of cytoplasmic mitochondrial dsDNA associated with mitochondrial dysfunction. Thus, silencing of STING avoids these negative consequences of LKB1 inactivation, while facilitating immune escape. SIGNIFICANCE: Oncogenic KRAS-mutant lung cancers remain treatment-refractory and are resistant to ICB in the setting of LKB1 loss. These results begin to uncover the key underlying mechanism and identify strategies to restore STING expression, with important therapeutic implications because mitochondrial dysfunction is an obligate component of this tumor subtype.See related commentary by Corte and Byers, p. 16.This article is highlighted in the In This Issue feature, p. 1.
©2018 American Association for Cancer Research.
Conflict of interest statement
Conflicts of interest
D.A.B. is a consultant for N of One, has received honoraria from Loxo Oncology and Madalon Consulting, and research grants from BMS and Novartis.
Figures


Comment in
-
Evading the STING: LKB1 Loss Leads to STING Silencing and Immune Escape in KRAS-Mutant Lung Cancers.Cancer Discov. 2019 Jan;9(1):16-18. doi: 10.1158/2159-8290.CD-18-1286. Cancer Discov. 2019. PMID: 30626603 Free PMC article.
References
-
- Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res 2016;76(5):999–1008 doi 10.1158/0008-5472.CAN-15-1439. - DOI - PMC - PubMed
-
- Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018. doi 10.1158/2159-8290.CD-18-0099. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

