Elucidation of the Amygdalin Pathway Reveals the Metabolic Basis of Bitter and Sweet Almonds (Prunus dulcis)
- PMID: 30297455
- PMCID: PMC6236625
- DOI: 10.1104/pp.18.00922
Elucidation of the Amygdalin Pathway Reveals the Metabolic Basis of Bitter and Sweet Almonds (Prunus dulcis)
Abstract
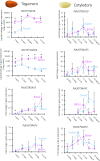
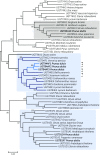
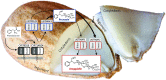
Almond (Prunus dulcis) is the principal Prunus species in which the consumed and thus commercially important part of the fruit is the kernel. As a result of continued selection, the vast majority of almonds have a nonbitter kernel. However, in the field, there are trees carrying bitter kernels, which are toxic to humans and, consequently, need to be removed. The toxicity of bitter almonds is caused by the accumulation of the cyanogenic diglucoside amygdalin, which releases toxic hydrogen cyanide upon hydrolysis. In this study, we identified and characterized the enzymes involved in the amygdalin biosynthetic pathway: PdCYP79D16 and PdCYP71AN24 as the cytochrome P450 (CYP) enzymes catalyzing phenylalanine-to-mandelonitrile conversion, PdUGT94AF3 as an additional monoglucosyl transferase (UGT) catalyzing prunasin formation, and PdUGT94AF1 and PdUGT94AF2 as the two enzymes catalyzing amygdalin formation from prunasin. This was accomplished by constructing a sequence database containing UGTs known, or predicted, to catalyze a β(1→6)-O-glycosylation reaction and a Basic Local Alignment Search Tool search of the draft version of the almond genome versus these sequences. Functional characterization of candidate genes was achieved by transient expression in Nicotiana benthamiana Reverse transcription quantitative polymerase chain reaction demonstrated that the expression of PdCYP79D16 and PdCYP71AN24 was not detectable or only reached minute levels in the sweet almond genotype during fruit development, while it was high and consistent in the bitter genotype. Therefore, the basis for the sweet kernel phenotype is a lack of expression of the genes encoding the two CYPs catalyzing the first steps in amygdalin biosynthesis.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures



Comment in
-
Setting and Diffusing the Cyanide Bomb in Plant Defense.Plant Physiol. 2018 Nov;178(3):956-957. doi: 10.1104/pp.18.01214. Plant Physiol. 2018. PMID: 30425157 Free PMC article. No abstract available.
References
-
- Andersen MD, Busk PK, Svendsen I, Møller BL (2000) Cytochromes P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J Biol Chem 275: 1966–1975 - PubMed
-
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA (1998a) Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol 36: 393–405 - PubMed
-
- Bak S, Nielsen HL, Halkier BA (1998b) The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol 38: 725–734 - PubMed
-
- Bak S, Olsen CE, Halkier BA, Møller BL (2000) Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol 123: 1437–1448 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

