Tuneable poration: host defense peptides as sequence probes for antimicrobial mechanisms
- PMID: 30297841
- PMCID: PMC6175903
- DOI: 10.1038/s41598-018-33289-y
Tuneable poration: host defense peptides as sequence probes for antimicrobial mechanisms
Erratum in
-
Author Correction: Tuneable poration: host defense peptides as sequence probes for antimicrobial mechanisms.Sci Rep. 2018 Nov 19;8(1):17266. doi: 10.1038/s41598-018-35521-1. Sci Rep. 2018. PMID: 30451943 Free PMC article.
Abstract
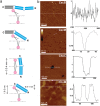
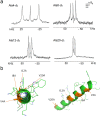
The spread of antimicrobial resistance stimulates discovery strategies that place emphasis on mechanisms circumventing the drawbacks of traditional antibiotics and on agents that hit multiple targets. Host defense peptides (HDPs) are promising candidates in this regard. Here we demonstrate that a given HDP sequence intrinsically encodes for tuneable mechanisms of membrane disruption. Using an archetypal HDP (cecropin B) we show that subtle structural alterations convert antimicrobial mechanisms from native carpet-like scenarios to poration and non-porating membrane exfoliation. Such distinct mechanisms, studied using low- and high-resolution spectroscopy, nanoscale imaging and molecular dynamics simulations, all maintain strong antimicrobial effects, albeit with diminished activity against pathogens resistant to HDPs. The strategy offers an effective search paradigm for the sequence probing of discrete antimicrobial mechanisms within a single HDP.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

