An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila
- PMID: 30297884
- PMCID: PMC6175926
- DOI: 10.1038/s41467-018-06537-y
An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila
Abstract
Being relatively simple and practical, Drosophila transgenic RNAi is the technique of top priority choice to quickly study genes with pleiotropic functions. However, drawbacks have emerged over time, such as high level of false positive and negative results. To overcome these shortcomings and increase efficiency, specificity and versatility, we develop a next generation transgenic RNAi system. With this system, the leaky expression of the basal promoter is significantly reduced, as well as the heterozygous ratio of transgenic RNAi flies. In addition, it has been first achieved to precisely and efficiently modulate highly expressed genes. Furthermore, we increase versatility which can simultaneously knock down multiple genes in one step. A case illustration is provided of how this system can be used to study the synthetic developmental effect of histone acetyltransferases. Finally, we have generated a collection of transgenic RNAi lines for those genes that are highly homologous to human disease genes.
Conflict of interest statement
The authors declare no competing interests.
Figures
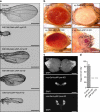
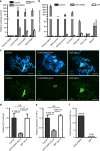
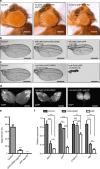


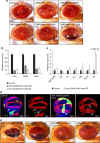
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

