Discovering Causal Relationships in Grapevine Expression Data to Expand Gene Networks. A Case Study: Four Networks Related to Climate Change
- PMID: 30298082
- PMCID: PMC6161569
- DOI: 10.3389/fpls.2018.01385
Discovering Causal Relationships in Grapevine Expression Data to Expand Gene Networks. A Case Study: Four Networks Related to Climate Change
Abstract
In recent years the scientific community has been heavily engaged in studying the grapevine response to climate change. Final goal is the identification of key genetic traits to be used in grapevine breeding and the setting of agronomic practices to improve climatic resilience. The increasing availability of transcriptomic studies, describing gene expression in many tissues and developmental, or treatment conditions, have allowed the implementation of gene expression compendia, which enclose a huge amount of information. The mining of transcriptomic data represents an effective approach to expand a known local gene network (LGN) by finding new related genes. We recently published a pipeline based on the iterative application of the PC-algorithm, named NES2RA, to expand gene networks in Escherichia coli and Arabidopsis thaliana. Here, we propose the application of this method to the grapevine transcriptomic compendium Vespucci, in order to expand four LGNs related to the grapevine response to climate change. Two networks are related to the secondary metabolic pathways for anthocyanin and stilbenoid synthesis, involved in the response to solar radiation, whereas the other two are signaling networks, related to the hormones abscisic acid and ethylene, possibly involved in the regulation of cell water balance and cuticle transpiration. The expansion networks produced by NES2RA algorithm have been evaluated by comparison with experimental data and biological knowledge on the identified genes showing fairly good consistency of the results. In addition, the algorithm was effective in retaining only the most significant interactions among the genes providing a useful framework for experimental validation. The application of the NES2RA to Vitis vinifera expression data by means of the BOINC-based implementation is available upon request (valter.cavecchia@cnr.it).
Keywords: ERF; NES2RA; Vitis vinifera; abscisic acid ABA; climate change; flavonoids; gene network; stilbenoids.
Figures
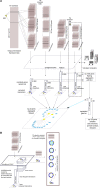
References
-
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. 10.1105/tpc.104.022897 - DOI - PMC - PubMed
-
- Alexa A., Rahnenfuehrer J. (2015). Gene Set Enrichment Analysis With topGO. Available at: https://www.bioconductor.org/packages/devel/bioc/vignettes/topGO/inst/do...
-
- Anderson D. P. (2004). “BOINC: a system for public-resource computing and storage,” in Proceedings of the 5th IEEE/ACM International Workshop on Grid Computing GRID ’04, (Washington, DC: IEEE Computer Society), 4–10. 10.1109/GRID.2004.14 - DOI
LinkOut - more resources
Full Text Sources

