Combining Developmental and Perturbation-Seq Uncovers Transcriptional Modules Orchestrating Neuronal Remodeling
- PMID: 30300589
- PMCID: PMC6179959
- DOI: 10.1016/j.devcel.2018.09.013
Combining Developmental and Perturbation-Seq Uncovers Transcriptional Modules Orchestrating Neuronal Remodeling
Abstract
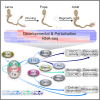
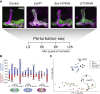
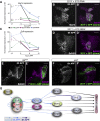
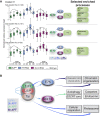
Developmental neuronal remodeling is an evolutionarily conserved mechanism required for precise wiring of nervous systems. Despite its fundamental role in neurodevelopment and proposed contribution to various neuropsychiatric disorders, the underlying mechanisms are largely unknown. Here, we uncover the fine temporal transcriptional landscape of Drosophila mushroom body γ neurons undergoing stereotypical remodeling. Our data reveal rapid and dramatic changes in the transcriptional landscape during development. Focusing on DNA binding proteins, we identify eleven that are required for remodeling. Furthermore, we sequence developing γ neurons perturbed for three key transcription factors required for pruning. We describe a hierarchical network featuring positive and negative feedback loops. Superimposing the perturbation-seq on the developmental expression atlas highlights a framework of transcriptional modules that together drive remodeling. Overall, this study provides a broad and detailed molecular insight into the complex regulatory dynamics of developmental remodeling and thus offers a pipeline to dissect developmental processes via RNA profiling.
Keywords: RNA-seq; axon pruning; developmental-seq; mushroom body; neuronal remodeling; γ-neurons.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Awasaki T., Tatsumi R., Takahashi K., Arai K., Nakanishi Y., Ueda R., Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855867. - PubMed
-
- Bai J., Uehara Y., Montell D.J. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. - PubMed
-
- Barkai N., Leibler S. Circadian clocks limited by noise. Nature. 2000;403:267–268. - PubMed
-
- Biggin M.D., Tjian R. Transcription factors and the control of Drosophila development. Trends Genet. 1989;5:377–383. - PubMed
-
- Bischof J., Björklund M., Furger E., Schertel C., Taipale J., Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

