Paired CRISPR/Cas9 Nickases Mediate Efficient Site-Specific Integration of F9 into rDNA Locus of Mouse ESCs
- PMID: 30301136
- PMCID: PMC6213315
- DOI: 10.3390/ijms19103035
Paired CRISPR/Cas9 Nickases Mediate Efficient Site-Specific Integration of F9 into rDNA Locus of Mouse ESCs
Abstract
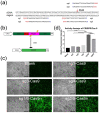
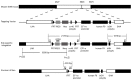
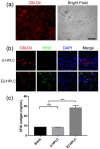
Hemophilia B (HB) is an X-linked recessive bleeding disorder, caused by F9 gene deficiency. Gene therapy combined with the CRISPR/Cas9 technology offers a potential cure for hemophilia B. Now the Cas9 nickase (Cas9n) shows a great advantage in reducing off-target effect compared with wild-type Cas9. In this study, we found that in the multicopy ribosomal DNA (rDNA) locus, the homology directed recombination (HDR) efficiency induced by sgRNA-Cas9n was much higher than sgRNA-Cas9, meanwhile without off-target in six predicted sites. After co-transfection into mESCs with sgRNA-Cas9n and a non-viral rDNA targeting vector pMrnF9, harboring the homology donor template and the human F9 expression cassette, a recombination efficiency of 66.7% was achieved and all targeted clones were confirmed to be site-specific integration of F9 in the rDNA locus by PCR and southern blotting. Targeted mESCs retained the main pluripotent properties and were then differentiated into hepatic progenitor like cells (HPLCs) and mature hepatocytes, which were characterized by hepatic markers and functional assays. Importantly, the differentiated cells could transcribe exogenous F9 and secrete coagulation factor IX (FIX) proteins, suggesting active transcription and stable inheritance of transgenes in the rDNA locus. After intrasplenical transplantation in severe combined immune deficiency (SCID) mice, targeted HPLCs could survive and migrate from spleen to liver, resulting in secretion of exogenous FIX into blood. In summary, we demonstrate an efficient and site-specific gene targeting strategy in rDNA locus for stem cell-based gene therapy for hemophilia B.
Keywords: CRISPR/Cas9 nickase; Hemophilia B; gene targeting; gene therapy; hepatic progenitor like cells; intrasplenic transplantation; mESCs; ribosomal DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J., Cuker A., Sullivan L.M., Majumdar S., Teitel J., et al. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N. Engl. J. Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

